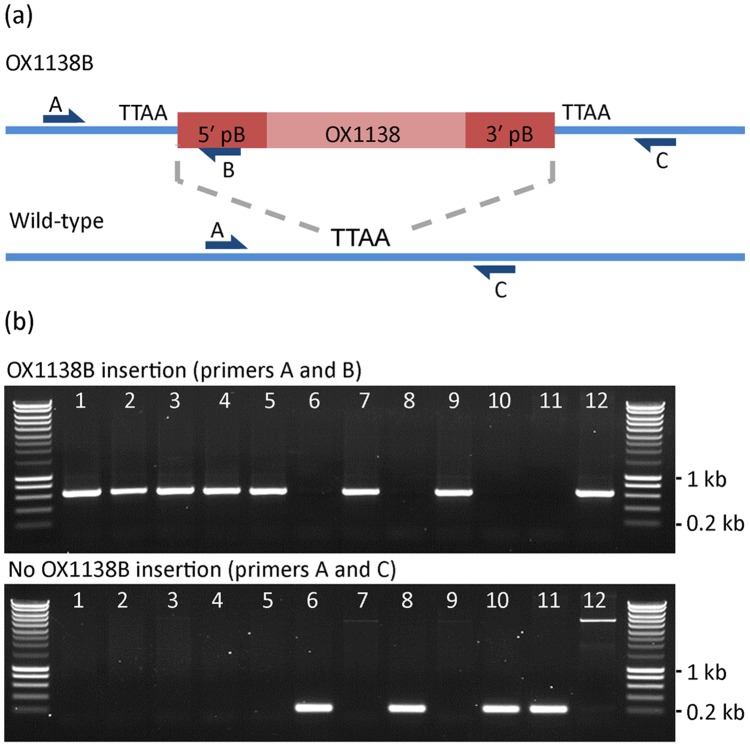
Figure 6. Genotyping moths by PCR.
(a) Schematic diagram showing genotyping PCR reactions for the OX1138B transgene insertion and its wild-type counterpart. The junction of the OX1138B insertion site in the wild-type sequence is indicated by the TTAA nucleotide sequence (piggyBac transposase recognition sequence which is duplicated on insertion of the piggyBac transposon). Primers and binding sites for primers A, B and C are indicated. PCR reactions containing primers A and B will amplify a fragment of 580 bp from OX1138B genomic DNA and no fragment from wild-type genomic DNA. PCR reactions containing primers A and C will amplify a fragment of 336 bp from wild-type genomic DNA and no fragment from OX1138B-homozygous genomic DNA. A moth that is heterozygous for the OX1138B insertion – carrying both the OX1138B and wild-type loci – would yield the amplified fragments in both PCR reactions. In OX1138B-positive genomic DNA, primers A and C would theoretically yield an amplified fragment, but the distance is usually too great for the PCR to amplify. (b) Gel images showing PCR genotyping results of moths from an experimental field cage trap (samples 1–10), a known wild-type moth (sample 11) and a known OX1138B-homozygous moth (sample 12). PCR for the transgene insertion yields the 580 bp fragment OX1138B moths (samples 1–5, 7 and 9) and PCR for the wild-type (no transgene insertion) locus yields the 336 bp fragment in APHIS moths (samples 6, 8 and 10). In the latter PCRs, amplification of the whole sequence spanning the transgene insertion is achieved, as seen in the reaction for sample 12.