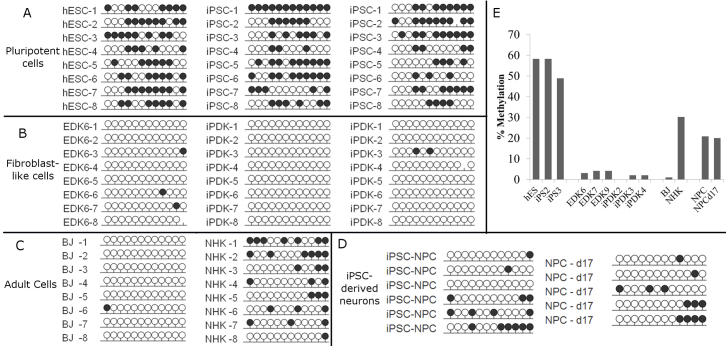
Fig. 6.
DNA methylation profile of the PDGFRβ gene promoter in pluripotent and pluripotent-derived cells. Bisulfite sequencing analysis of the PDGFRβ gene promoter was performed on ESC, iPSC, EDK, iPDK, primary fibroblasts (BJ) and keratinocyte (NHK) cell types, as well as iPSC-derived neural progenitors (iPSC-NPCs) and neurons following 18 days of differentiation (NPC-d18). Each dot represents one CpG site in the promoter region of PDGFRβ and each row represents a clonal isolate of DNA fragments from indicated cell types. Methylated CpGs are indicated by a black circle, and unmethylated CpGs are indicated by a white circle. (A) PDGFRβ is methylated in pluripotent cell types h9-hES, iPS2 and iPS3. (B) PDGFRβ is unmethylated in hES-derived EDK, iPS2-derived iPDK and iPS3-derived iPDK. (C) In adult cell types, PDGFRB promoter was unmethylated in BJ fibroblasts, compared with primary keratinocytes (NHK), which are relatively highly methylated. (D) iPSC-derived neural progenitors (NPCs) and neurons following 18 days of differentiation (NPC d18), demonstrated an intermediate level of methylation at the PDGFRβ promoter. (E) Quantification of bisulfite sequencing results showed greater than 50% methylation in pluripotent cell types, and 0–5% methylation in pluripotent-derived EDK and iPDK cells, and adult fibroblasts, but not neurons derived from iPSCs.