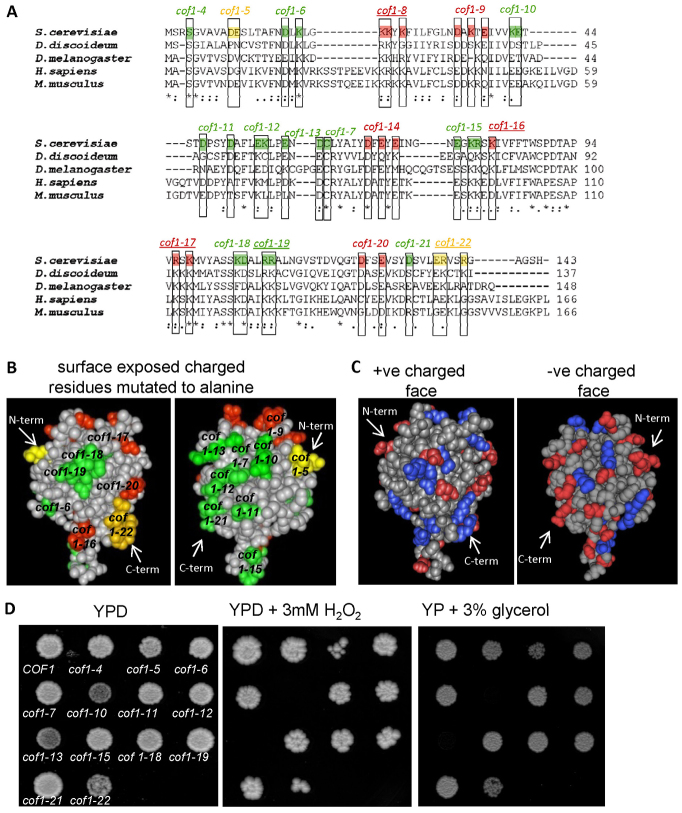
Fig. 1.
Analysis of recognised actin and non-actin regulatory surfaces of cofilin family proteins. Alignment of yeast cofilin (Saccharomyces cerevisiae NCBI accession; AAA13256.1), slime mould cofilin (Dictyostelium discoideum; NCBI accession BAA07198.1), twinstar from fruit fly (Drosophila melanogaster; NCBI accession NP_477034.1), human cofilin (Homo sapiens; NCBI accession CAA64685.1) and mouse cofilin (Mus musculus; Accession: AAA37433.1). Residues essential for actin binding and for viability in yeast are coloured red. Those that affect actin dynamics but which are not essential in yeast are coloured in yellow and those that do not affect yeast growth are labelled in green. The names of cofilin mutant alleles that impair normal PtdIns(4,5)P2 binding abilities of cofilin are underlined. (A) Space-filling models of the 3.00 Å yeast cofilin crystal structure (Protein databank reference 1QPV) with residues involved in actin binding (red and yellow) and not involved in actin regulation (green) highlighted. (B) Surface charged residues are depicted in red (−ve) and blue (+ve) on the yeast cofilin space-filling model. (C) Wild-type (COF1) and cofilin mutant strains were grown as colonies on agar plates containing YPD, YPD + 3 mM H2O2 or YP + 3% glycerol for 3 days (D).