Abstract
A tremendous wealth of data is accumulating on the variety and distribution of transposable elements (TEs) in natural populations. There is little doubt that TEs provide new genetic variation on a scale, and with a degree of sophistication, previously unimagined. There are many examples of mutations and other types of genetic variation associated with the activity of mobile elements. Mutant phenotypes range from subtle changes in tissue specificity to dramatic alterations in the development and organization of tissues and organs. Such changes can occur because of insertions in coding regions, but the more sophisticated TE-mediated changes are more often the result of insertions into 5′ flanking regions and introns. Here, TE-induced variation is viewed from three evolutionary perspectives that are not mutually exclusive. First, variation resulting from the intrinsic parasitic nature of TE activity is examined. Second, we describe possible coadaptations between elements and their hosts that appear to have evolved because of selection to reduce the deleterious effects of new insertions on host fitness. Finally, some possible cases are explored in which the capacity of TEs to generate variation has been exploited by their hosts. The number of well documented cases in which element sequences appear to confer useful traits on the host, although small, is growing rapidly.
The book whose publication we are celebrating in this colloquium indicates that Theodosius Dobzhansky had a very special interest in gene mutation and its causes. Dobzhansky recognized mutation as the “raw material” on which natural selection acts and as the first of three steps necessary for evolution to take place. However, the discovery of transposable elements (TEs) in the 1940s by Barbara McClintock occurred a decade later, and it was a further 30 years before the significance of her findings started to be fully appreciated. Sixty years ago, Dobzhansky was well aware of the mutagenic properties of ionizing radiation discovered in 1927 by H. J. Muller but acknowledged that much less than 1% of spontaneous mutations were attributable to this cause. He distinguished between spontaneous and induced mutations: “The former are those which arise in strains not consciously exposed to known or suspected mutation-producing agents.” He also pointed out that “since the name spontaneous constitutes only a thinly-veiled [sic] admission of the ignorance of the phenomenon to which it is applied, the quest for the causes of mutation has always occupied the attention of geneticists.” Although at that time no clues to its nature were yet available, Dobzhansky realized that a major piece of the mutation puzzle was still missing. We believe he would have been intrigued with the discoveries of TEs in natural populations that have taken place during the last 20 years and that he would have been an active participant in the continuing debate about their role in evolution.
Distribution and Classification
TEs are discrete segments of DNA that are distinguished by their ability to move and replicate within genomes. Since their discovery by Barbara McClintock ≈50 years ago (1), TEs have been found to be ubiquitous in most living organisms. They comprise a major component of the middle repetitive DNA of genomes of animals and plants. They are present in copy numbers ranging from just a few elements to tens, or hundreds, of thousands per genome. In the latter case, they can represent a major fraction of the genome, especially in some plants. For example, TEs recently have been estimated to make up >50% of the maize genome (2). In Drosophila, ≈10–15% of the genome is estimated to be made up of TEs, most of which are found in distinct regions of centric heterochromatin (3).
TEs are classified in families according to their sequence similarity. Two major classes are distinguished by their differing modes of transposition (4). Class I elements are retroelements that use reverse transcriptase to transpose by means of an RNA intermediate. They include long terminal repeat retrotransposons and long and short interspersed elements (LINES and SINES, respectively). Long terminal repeat retrotransposons are closely related to other retroelements of major interest, such as retroviruses (5). The gypsy element in Drosophila is an example of a rare type of retrotransposon that can sometimes also behave as a retrovirus (6).
Class II elements transpose directly from DNA to DNA and include transposons such as the Activator-Dissociation (Ac-Ds) family in maize, the Tam element in Antirrhinum, the P element in Drosophila, and the Tc1 element in the worm, Caenhorabditis elegans. Recently. a category of TEs has been discovered (7) whose transposition mechanism is not yet known. These miniature inverted-repeat TE (MITEs) have some properties of both class I and II elements. They are short (100–400 bp in length), and none so far has been found to have any coding potential. They are present in high copy number (3,000–10,000) per genome and have target site preference for TAA or TA in plants. MITEs such as the Tourist element in maize and the Stowaway element in Sorghum (7) are found frequently in the 5′ and 3′ noncoding regions of genes and are frequently associated with the regulatory regions of genes of diverse flowering plants. TEs with similar properties also have been described in Xenopus (8), humans (9, 10), and the yellow fever mosquito, Aedes aegypti (11).
Most, but not all, TE families are made up of both autonomous and nonautonomous elements. Whereas autonomous elements code for their own transposition, nonautonomous elements lack this ability and usually depend on autonomous elements from the same, or a different, family to provide a reverse transcriptase or transposase in trans.
This paper aims first to provide a brief, general description of the types of genetic variation caused by TEs in animals and plants and then to examine this variation within an evolutionary framework: (i) direct selection on TEs at the level of the DNA sequence (parasitic DNA); (ii) coevolution of TEs and their animal and plant hosts to avoid or mitigate the deleterious effects of insertion; and (iii) positive selection on elements that have evolved to provide some positive benefit to their hosts in addition to simply minimizing the harm they do.
Types of TE-Induced Genetic Variation
Like new mutations produced by any mutator mechanism, the majority of new TE-induced mutations are expected to be deleterious to their hosts. Those mutations that survive over long periods of evolutionary time are expected to be a small subsample of newly induced mutations. The property that distinguishes TE-induced mutations from those produced by other mutational mechanisms is their remarkable diversity and the degree to which their induction is regulated by both the host and the TE itself.
The genetic variability resulting from TEs ranges from changes in the size and arrangement of whole genomes to changes in single nucleotides. It may produce major effects on phenotypic traits or small silent changes detectable only at the DNA sequence level. It is important to note that TEs produce their mutagenic effects not simply on initial insertion into host DNA. TEs may also produce mutations when they excise, leaving either no identifying sequence or only small “footprints” of their previous presence. In addition, some TE-induced mutations that may be of evolutionary significance to their hosts, such as mutations in regulatory sequences (12), may take long periods of time to evolve new functions or these new functions may have been acquired a long time ago. Consequently, they may have lost their original identification as TEs. For these reasons, the reliance solely on the distribution of TE sequences in the genomes of contemporary species of animals and plants to deduce the long term evolutionary importance of TEs may produce a biased result that may not adequately reflect TE-associated events that occurred long in the past.
Deleterious effects of TEs can result not only from mutations caused by the insertion or excision of these elements at a single chromosomal site but also from genomic-level disruptive effects associated with TE transposition. For example, massive chromosome breakage in larval cells resulting from excision and transposition of genomic P elements has been implicated as the cause of temperature-dependent pupal lethality and sterility in hybrid dysgenesis in Drosophila melanogaster (13, 14).
A brief description of the types of genetic variability caused by TE activity follows, based largely on the types of host DNA involved. Some of the mutations described were generated in the laboratory and have been subjected to artificial selection under unnatural and noncompetitive conditions. Although these are generally not the class of mutations that are of interest from an evolutionary perspective, we include them here to provide some indication of the potentially wide spectrum of phenotypic changes associated with TE activity.
Insertions of TEs into Exons of Host Genes.
On average, TEs that insert within the exons of genes are most likely to result in null mutations because of the sensitivity of these regions to frame shift mutations and the lack of tolerance of highly conserved regions to most mutations of any kind. However, those mutations that are not simply inviable can provide interesting and sometimes spectacular phenotypic variability. In Drosophila, a series of null alleles at the X-linked, white locus allowed the first identification of the P element in D. melanogaster as the causal agent of P–M hybrid dysgenesis (15). The insertion of both the P element and the copia element into exon sequences interrupted the coding sequences and the production of the red eye pigment by the wild-type gene. The result is a bleached white eye phenotype that reflects the lack of pigmentation. Such a null mutation can be maintained in the laboratory but is unlikely to survive in natural populations.
A good classic example is the insertion of an element of the Ac-Ds family into wx-m9, an allele of the waxy locus in maize first discovered by McClintock (16). The mutation is caused by the insertion of Ds (Dissociator) into the 10th exon of the waxy locus. This was the first element to be cloned from maize, and it is of continuing interest because it is spliced, resulting in partial revertant activity (17, 18). In this case, the effect of the insertion is attenuated by the loss through splicing of the TE after transcription.
Insertions into Regulatory Regions of Genes.
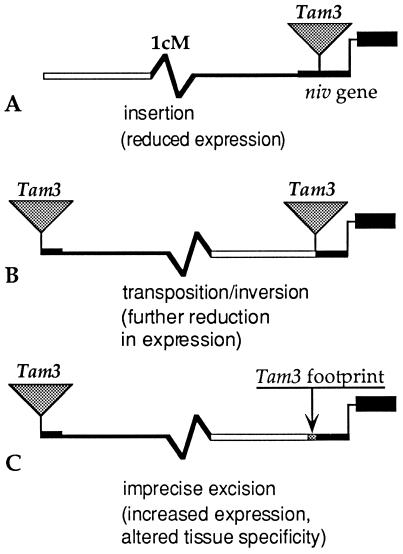
An excellent example of this type is the insertion of gypsy into the 5′ upstream region of the yellow gene in Drosophila, which causes a loss of expression of the yellow gene in specific tissues (19). The loss of expression in some tissues and not others in this case is the result of the interaction of the element, tissue-specific enhancers upstream of the element, and specific host factors. In Antirrhinum, a Tam3 element was observed to insert into a region 5′ of the niv gene, which is involved in the synthesis of anthocyanin pigments. The initial insertion was observed to down-regulate expression of the gene. However, a series of rearrangements mediated by this element resulted in a change in the level and tissue specificity of expression of niv (Fig. 1). The net effect is a new and novel distribution of anthocyanin pigment in the flower tube (20). This series of mutations exemplifies the potential for TE-mediated “rewiring” of regulatory networks, in this case by bringing new regulatory sequences in proximity to exonic sequences via an inversion, followed by an imprecise excision event.
Figure 1.
Rearrangements associated with TE activity near a gene result in altered tissue specificity. (A) In the original isolate, a Tam3 element had inserted 64 bp upstream of the start of transcription of the niv gene of Antirrhinum. The result of this insertion was a reduced level of expression. (B) A derivative of the initial insertion allele carries an inversion flanked by two copies of the transposon. This allele confers an additional reduction in the level of expression. (C) Excision of the element closest to the niv gene left a short (26 bp) “footprint.” This rearrangement resulted in an increase in the level of niv gene expression as well as a novel pattern of expression, presumably due to the juxtaposition of a novel sequence with the niv gene TATA box and coding sequences [adapted from Lister et al. (20)].
TE activity can result in even more complex rearrangements that can have effects on gene regulation. In maize, the insertion of a Mu (Mutator) element into the TATA box of Adh1 changes the tissue specificity of RNA expression (21). Of interest, excision of this element caused a complex series of duplications and inversions whose net effect was to cause additional changes in tissue specificity (22). Kloeckener–Gruissem and Freeling (22) suggest that this kind of “promoter scrambling” may represent a more general process by which transposons produce variants of a type not produced by other mechanisms. Furthermore, in this case, like that of the Tam element in the niv gene of Antirrhinum, the TE footprint left behind after the element has generated the new mutation is small enough to be invisible to TE probes.
Insertions in Introns.
TEs that insert into introns generally have a greater chance to survive because these insertions are less visible to natural selection. Many of them are probably successfully spliced out during mRNA processing and have no obvious effect on the function of the gene. Even when spliced, however, introns are sometimes the site of regulatory sequences. In these cases, TE insertions into introns can affect gene regulation in surprising ways. For instance, the insertion of Mu elements into an intron of the Knotted locus in maize induces ectopic expression of the gene, suggesting that the intron carries sequences normally required to repress expression of the gene in certain tissues (23). Similarly, in Antirrhinum, complementary floral homeotic phenotypes result from opposite orientations of a Tam3 transposon in an intron of the ple gene (24) .
Insertions in Heterochromatin.
Middle repetitive DNA sequences, including TEs, are an important component of β heterochromatin in Drosophila, and retrotransposons constitute a considerable fraction of this DNA (25). Recent work by Pimpinelli and coworkers (3) has revealed that TE clustering into discrete regions of heterochromatin is a general property of elements in Drosophila. The cause of this distribution pattern is an open question. In some cases, a heterochromatic location probably reduces the probability of elimination of inserted sequences from the genome by ectopic recombination, the mechanism believed to be largely responsible for controlling TE copy number in chromosome arms (26). However, some centric heterochromatic regions have been described as a graveyard for dead elements, rather than a safe haven for active elements, because the majority found there appear to be inactive and highly diverged sequences. For example, LINE-like I elements cloned from the Charolles-reactive strain of D. melanogaster contain no active euchromatic I factors, only defective copies that are embedded in clusters of defective copies of other retroelements (27). In contrast, elements such as mdg1 have been found to have a nested arrangement within other retrotransposons located in euchromatic chromosome regions (28, 29). These sites appear to exhibit properties of intercalary heterochromatin (25) and may be responsible for the properties of ectopic pairing, susceptibility to breakage, and late replication that are characteristic of this type of chromatin.
Mediation of Recombination.
TE-mediated increases in the rate of recombination have consequences not only for genetic variation at individual loci. This recombination activity can also result in more general changes in both fine and gross structural characteristics of chromosomes. For instance, analysis of mutations in the mei-41 and mus302 genes required for normal postreplication repair in D. melanogaster (30) revealed a striking stimulation of site-specific gene conversion and recombination mediated by P element transposition. As a consequence of the selection against the negative effects of ectopic recombination, this is postulated to be the mechanism chiefly responsible for the removal of certain subsets of TEs from genomes and a means for controlling copy number (26, 31). It is worth noting, however, that not all rearrangements caused by ectopic recombination are necessarily selected against. From an evolutionary perspective, rare surviving chromosomal rearrangements could be of significance. For example, Lyttle and Haymer (32) demonstrated the presence of the hobo element at the breakpoints of several endemic, but not cosmopolitan, inversions in D. melanogaster natural populations from Hawaii. This result is consistent with the recent introduction of hobo elements to D. melanogaster by horizontal transfer (33) and the subsequent production of these inversions as a consequence of the activity of these elements.
Effects on Quantitative Variability.
The P element in Drosophila provides one of the most compelling demonstrations of TE-induced genetic variability in quantitative genetic traits. A series of experiments (34, 35) has shown that quantitative variability for bristle number is induced by P–M hybrid dysgenesis and is demonstrable by directional artificial selection. Furthermore, in well controlled experiments (36), a dramatic increase in new additive genetic variation in abdominal bristle number was observed that was 30 times greater than that expected from spontaneous mutation. In another Drosophila study (37), an excess of P element mutations having large effects on metabolic characters was observed relative to those expected. Significant among-line heterogeneity indicated that the mutational target site for enzyme activity is large and that most of the mutations must be regulatory. It was concluded that the large pleiotropic effects observed had important consequences for metabolic characters.
Evolutionary Considerations
The idea that TEs are primarily parasitic is not at all inconsistent with a role for these elements in the evolution of their hosts. Indeed, as documented below and elsewhere (12, 38–40), there is a growing body of evidence for coadaptation by both elements and their hosts to the long term presence in the genome of these parasitic sequences. In some cases, it appears that this coadaptation even may have lead to the use of TE sequences for essential and beneficial host functions. In this section, we explore all three perspectives.
TEs as Genomic Parasites
The intrinsically parasitic nature of active TEs (41–43) accounts for their undisputed ability to invade new species, increase in copy number, and survive over long periods of evolutionary time. The replicative advantage of TEs (44) is responsible for this ability, which is facilitated by their generally compact structure and inclusion of the coding capacity for transposition within their sequences. Natural selection acting on TEs at the level of the DNA sequence is responsible for maintaining their essentially parasitic properties. For example, P elements can rapidly invade a naive population of D. melanogaster, despite extremely strong negative selection at the host level in the form of high frequencies of temperature-dependent gonadal sterility (45).
A proclivity for horizontal transfer is consistent with the role of TEs as genomic parasites. The life cycle of TEs in any single phylogenetic lineage can apparently last for many thousands or millions of years and can be considered as a succession of three phases: dynamic replication, inactivation, and degradation (46, 47). The transposition of both major classes of elements is error-prone and produces nonautonomous elements that often repress the transposition rate of active elements. Over long periods of evolutionary time, there is a tendency for a family of elements to degrade in coding capacity, but horizontal transfer to another host lineage provides the opportunity for active TEs to move to another lineage and begin the cycle over again (46, 48, 49). There is evidence that TEs do transfer horizontally more frequently than nonmobile genes (46). Class II elements such as mariner and P elements provide good examples of TE horizontal transfer (50–52), but a major puzzle remains regarding the mechanism by which horizontal transfer is achieved (49).
Coadaptations to Mitigate Reduced Host Fitness
Like viruses, TEs are dependent on their host organisms for survival, but unlike viruses, most TEs do not have a phase in their life cycle in which they can survive independent of their hosts. Therefore, coevolution and coadaptation of TEs with host genomes is expected to play a particularly important role in the long term survival of these element families. Given that the majority of new insertions tend to be deleterious to hosts, it is in the interests of both parties to mitigate or remove such deleterious effects. TEs can insert in many locations other than exons. In these noncoding sequences, they are likely to increase their probability of survival because of less visibility to natural selection. Examples of some of the ways that coadaptations by both mobile elements and their hosts appear to have evolved are described below and are summarized in Table 1.
Table 1.
Possible coevolved mechanisms to mitigate reduction in host fitness
| Mechanism | Examples |
|---|---|
| Insertion bias for noncoding regions | Preferential insertion in regulatory regions (1); nested retrotransposons in maize (2); clustered I elements (27); mdg1 in Drosophila (28, 29) |
| Pre-mRNA splicing | Splicing from the maize wx gene (18); various genes in Drosophila (57), C. elegans (59), and plants (58) |
| Tissue specificity of transposition | Repression of P element transposition in somatic cells at the level of RNA processing (60) |
| TE copy number regulation | Methylation of maize Ac, Spm, and Mu by host factors (63, 64); type I and type II P element-encoded repressors (13, 68, 70–72); Ac dosage effects (16); Spm repressor action (73–75) |
| Regulators of mutant phenotype expression | Transposase-dependent expression of mutant phenotype caused by insertion of Spm in maize (73); masking of mutant phenotypes by alleles of host suppressor genes in Drosophila (40) |
Insertion Bias for Noncoding Regions.
A dramatic demonstration of the preference for new P element insertions into potentially regulatory regions of genes, rather than exons, is provided by P elements in Drosophila (53). Only a small minority of P elements was observed to have inserted in coding sequences, and these elements were in the 5′ portion. Thus, there is a strong bias in favor of insertion in the 5′ end of the gene and especially for the 5′ untranslated region. Because these insertions all caused an obvious mutant phenotype (they failed to compliment a deficiency), this result is actually an understatement of the true number of insertions into or near regions of genes often involved in regulation. This preference is supported by the remarkable observation that, in another experiment, >65% of the 500 independent P element enhancer trap insertions were expressed in a spatially and temporally restricted fashion (54) .
Another good example of preferential insertion of elements outside of coding regions is provided in yeast (55). Of over 100 new insertions of Ty1 observed in chromosome III, nearly all were inserted into or near either tRNA genes or preexisting long terminal repeats; only 3% were found in ORFs. Distribution patterns favoring the 5′ noncoding sequences of genes were observed for MITE elements in plants (7) and animals (e.g., see ref. 11). However, it is not clear whether this pattern represents an insertion preference or whether it results from strong earlier selection against insertions into other regions. Similarly, in view of the large number of group II and group III introns present in the chloroplast genome of E. gracilis, the complete absence of introns in rRNA and tRNA genes is striking (56). One possibility is that secondary structure features of rRNA and intron RNA or tRNA and intron RNA (if they were present in the same pre-mRNA molecule) would interact in such a way as to prevent one or both from functioning.
Preference for Insertion into Preexisting Elements.
In some cases, it is clear that unrestricted transposition would be absolutely disastrous for the host. As mentioned above, a remarkable number of retroelements, probably representing well over 50% of the genome, are found between genes in maize (2). The five most abundant families make up more than 25% of the genome, and one family alone, Opie, makes up 10–15% of the maize genome. Despite this ubiquity, few homologies in the database were found among maize genes; these elements appeared to have a pronounced preference for regions outside of genes. Indeed, fully half of the elements examined were found nested within other elements.
Splicing from Pre-mRNA Transcripts.
It has been suggested that splicing of TEs from exonic sequences has evolved as a means by which TEs can minimize their deleterious host impact. We refer back to the waxy mutation described earlier (18). In that case, splicing of the Ds element inserted into the waxy gene results in partial reversion of the mutation caused by the original insertion. Presumably, even a partial amelioration of the mutant phenotype can provide some selective advantage to those TEs capable of providing it. Additional examples are found in Drosophila, C. elegans, and other plants (57–59). In addition, Marillonnet and Wessler have observed tissue-specific splicing of an element (S. Wessler, personal communication), suggesting that TEs could potentially play a role in the evolution of tissue-specific regulation of some genes. These examples may be illustrating an evolutionary spectrum, from purely parasitic behavior to functional significance for the host. The original capacity to be spliced may have arisen as a way to minimize the impact of insertions into coding sequences but on the road from poorly spliced variants that simply ameliorate mutant phenotypes to fully effective and selectively invisible splicing may have come the opportunity to develop new classes of regulation, such as tissue specificity.
Tissue Specificity of TE Activity.
A good example of a likely adaptation of a TE to its host is the restriction of transposition of P and I elements to the germ line (14). It is of mutual benefit for an element to transpose in those tissues that will ensure transmission to the next host generation, but to curtail activity in somatic tissues is likely to result in loss of host fitness without providing any benefit to the transposon. Repression of P element transposition in somatic cells occurs on the level of RNA processing (60). The 2–3 intron is spliced only in the germ cells, resulting in the absence of transposase in somatic cells. Splicing of this intron is prevented in the somatic cells by an 87-kDa protein that binds to a site in exon 2 located 12–31 bases from the 5′ splice site (61). An existing host-splicing mechanism apparently has been coopted for this purpose (61, 62) that has been highly conserved during evolution.
Host Regulation of TE Copy Number.
Good examples of host regulation of copy number are found in maize and Drosophila. Unknown host-encoded factors specifically methylate Ac, Spm, and Mutator elements in maize (63, 64). With Mu (64), the example is particularly striking because it represents the global methylation of dozens of previously active elements simultaneously in a single generation. The methylation is not simply related to structural features of insertion sites of Mu elements because they become specifically methylated even when inside of genes; modification is rarely detected in the flanking sequences within the genes (65) .
In D. melanogaster females, expression of the gypsy element envelope gene is strongly repressed by one copy of the nonpermissive allele of flamenco [reviewed by Bucheton (6)]. A less dramatic reduction in the accumulation of other transcripts and retrotranscripts also is observed. These effects correlate well with the inhibition of gypsy transposition in the progeny of these females and are therefore likely to be responsible for this phenomenon. The effects of flamenco on gypsy expression apparently are restricted to the somatic follicle cells that surround the maternal germline.
Self-Regulation of TE Copy Number.
It has been shown theoretically that self-regulation of TEs cannot evolve if it is assumed that deleterious effects on host fitness are caused by increased copy number alone or are not caused by dominant lethals (66). However, if the deleterious effects are immediate and occur as a direct consequence of transposition itself, then there may be a selective advantage to elements with reduced transposition rates that still allow them to spread in the genome but at a reduced cost to their host [reviewed by Brookfield (67)].
The activity of the P elements in D. melanogaster is regulated by element-encoded repressor products. These repressors fall into two discrete categories, type I and type II. Type I repressors are responsible for a cellular condition known as P cytotype, which depends on a 66-kDa, P element-encoded, repressor of transposition and excision (68). The genomic position of repressor elements determines the maternal vs. zygotic inheritance of P cytotype (69). Type II repressors usually have large internal deletions, are sensitive to genomic location, but show no maternal inheritance (13, 70–72) .
In plants, the Ac element shows dosage effects; an increase in number of elements results in a decreased number of transpositions of the element (16). This could be interpreted as a response of the plant to increases in the level of Ac transposase or as an autoregulatory mechanism. Similarly, Spm:tnpA can protect Spm from methylation but may also act as a repressor of Spm (73, 74). Additionally, some deleted Spm elements can repress full length Spm elements in trans (75) .
Regulators of Expression of Mutant Phenotypes.
Very early in the investigation of mutable alleles in maize, it was discovered that the expression of some alleles depended on the presence or absence of a second factor (76). In these cases, that factor was the source of the transposase. However, as is clear from the example of some gypsy insertions whose mutant phenotype is only manifested in the presence of both Su(Hw) and a second host encoded factor, mod(mdg4) (77), TE-induced mutant alleles can also become ameliorated by other factors as well.
The mutant phenotypes associated with many retrotransposon insertions are masked by alleles of host suppressor genes that act as trans-regulators of retrotransposon expression (40). The argument made is that such suppressor action may allow insertion mutations to partially, or completely, escape the action of purifying selection and allow them to persist or even increase in frequency in natural populations. There is evidence for the presence of host genes with suppressor function in natural populations of Drosophila (78).
TE-Induced Characters Having Benefit to the Host
There has been considerable debate whether, in addition to deleterious effects on fitness, TE-induced variability has any significance for host organisms over evolutionary time (40). The generally unpredictable nature of TE movements, coupled with the paucity of fixed insertion sites for TEs in species such as Drosophila (26), has lead some to reject the possibility of TEs having any significant evolutionary importance, other than as molecular parasites. However, there is a rapidly growing list of possible examples of TEs having evolved highly sophisticated functions, as shown by the examples briefly described below and summarized in Table 2.
Table 2.
Examples of TEs having functions that benefit their hosts
| New function | Examples |
|---|---|
| Insertions with regulatory functions | More than 20 examples of insertions into regulatory regions of genes (12, 38, 39) |
| “Molecular domestication” | P element tandem repeats in D. obscura group may provide a new host gene function (89) |
| Source of new introns | Introns and twintrons in Euglena gracilis plastids (24) |
| Replacement of normal host functions | Repair of damaged chromosome ends by HET-A and TART in Drosophila (92, 93) |
| A role in host cell repair mechanisms | Endogenous retroelements associated with repair of DSBs in yeast (94, 95) |
| Mediation of concerted evolution | P element-mediated changes in subtelomeric repeat numbers in D. melanogaster (96) |
| Possible functions of heterochromatic TE clusters | Developmentally programmed changes in DNA content; expression of heterochromatically embedded loci; genomic housekeeping functions (25) |
DSB, double-strand chromosome break.
Insertions with Host Gene Regulatory Functions.
It has been speculated for some time that changes in cis-regulatory regions of duplicated genes may be more important for the evolution and divergence of functional and morphological characters than mutations in coding sequences (see, e.g., refs. 79 and 80). However, only recently has evidence started to accumulate to support this hypothesis. In Drosophila, for instance, the three homeotic genes paired (prd), gooseberry (gsb), and gooseberry neuro (gsbn) have evolved from a single ancestral gene, following gene duplication. They now have distinct developmental functions during embryogenesis. The three corresponding proteins PRD, GSB and GSBN are transcription factors. Li and Noll (81) demonstrated that the three proteins are interchangeable with respect to their regulatory functions and that their distinct developmental functions are a consequence of changes in the regulatory sequences rather than in the proteins themselves. Because they lend themselves so well to changes in the architecture of promoter regions, it is likely that TE mutations have been involved in this kind of regulatory evolution (82).
The potential importance of TEs as modifiers of the expression of normal plant genes has been highlighted by recent findings in plants. Long terminal repeat retrotransposons and MITEs have been found to be associated with the genes of many plants where some of these TEs contribute regulatory sequences (7). Furthermore, the MITE elements recently discovered in Aedes aegypti (11) also are associated closely with genes. In domesticated rice, Oryza sativa, a computer-based search revealed 32 common sequences belonging to nine putative mobile element families (83). Four of these families had been previously described, but five families were first discovered through this computer search, and four of these five had characteristics of MITES.
New Patterns of Tissue-Specific Expression.
TEs can contribute to the functional diversification of genes by supplying cis-regulatory domains altering expression patterns. Earlier, we described the insertion of the gypsy element into a 5′ upstream region of the yellow gene in Drosophila causing a loss of expression of this gene in specific tissues (84). The tissue-specific alterations in expression (a kind of mutation that is more subtle than simply knocking out a gene) is due to the presence of a specific sequence of DNA that is bound by Su(Hw), which is thought to be a transcription factor. More interesting, the Su(Hw) binding sequence seems to act as a general “buffer” that helps to define structural domains in the chromatin (77). Thus, gypsy may serve to introduce domains of regulation into given regions of the chromosome. This may have arisen initially as a means by which the TE could buffer itself from its chromosomal environment, but this kind of domain alteration could certainly also result in interesting variations in gene regulation as well.
In addition to simply buffering chromosomal regions, many TEs are specifically expressed only in particular tissues at particular times. Based on recent findings, it appears that tissue specificity is a general feature of all retrotransposons in Drosophila. The expression patterns of 15 different families of long terminal repeat-containing retrotransposons were examined by Ding and Lipshitz (85) during normal development in different wild-type strains of D. melanogaster. Each family exhibited a pattern typical of spatial and temporal expression during embryogenesis, suggesting that each TE harbors cis-regulatory factors that interact specifically with host transcription factors. These mobile cis-regulatory factors could potentially act to modify the expression of any number of host genes.
Other Types of Insertions with Regulatory Functions.
Some of the examples given thus far are anecdotal; they represent laboratory observations as to the kinds of changes that TEs can introduce into the host genome, rather than changes that have actually contributed to the evolution of the host. However, Britten (12, 38, 39) has used stringent criteria for the identification of strong cases of the involvement of TEs in the actual evolution of gene regulation. He maintains that a long term perspective is necessary in identifying and understanding mutations important for gene regulation. The number of cases he has identified is small, but growing. In addition to the plant MITE examples discussed above, he includes cases involving Alu-containing, T cell-specific enhancers in the human CD8a gene (86), the association of a retrovirus-related element with androgen regulation of the sex-limited protein (Slp) gene in mouse (87), and inverted repeats in the CyIIIa actin gene of sea urchin (88).
Tandem Repeats of P-Related Sequences in Drosophila.
A number of tandem P element repeats in three closely related species of the obscura group provides a very interesting example of several unrelated TE sequences evolving together that may provide a type of host gene function, which Miller et al. have termed “molecular domestication” (89). In this case, the P elements have lost all of their terminal repeats and thus can no longer transpose. Remarkably, each cluster unit consists of a cis-regulating section composed of insertion sequences derived from unrelated TEs, followed by the first three exons that, in mobile P elements, code for a 66-kDa protein that represses P element transposition. In contrast to this normal repressor function, these stationary P element repeats are hypothesized to have evolved the function of transcription factors (90).
A Source of New Introns.
Some retroelements are apparently fully adapted to their niche within exonic sequences. For example, the 143-kb Euglena gracilis plastid genome contains 155 group II and group III introns (56), nearly 10 times the number in any other known plastid DNA. The original introns were likely mobile, retrotransposable genetic elements that invaded the genome from another organism, relying in part on internally encoded enzyme activities for mobility. The group III introns appear to be streamlined versions of group II introns, sharing a common evolutionary ancestor with a group II intron. Among the E. gracilis introns are a number of introns-within-introns (twintrons), suggesting that these elements themselves have been targets of intron insertions. In one particularly interesting example (91), a group III intron is formed from domains of two individual group II introns. The authors suggest the possibility that “the introduction of one catalytic RNA into a functional domain of another catalytic RNA, through a process similar to twintron formation, can result in new combinations of sequences and structural domains that might lead to new RNA catalyzed reactions significant for RNA evolution.”
Telomeres in Drosophila.
An unusually finely tuned system between the host genome and mobile elements has evolved in Drosophila to take over a basic cellular function. Several retroelements, such as HET-A and TART, carry out the function of replacing damaged chromosome ends that is performed by telomerase in other insects (92, 93). The insertion frequency of the TEs involved has become adapted to match the average rate of telomere loss to maintain constant chromosome size. This is the best example to date of a TE providing a vital function to its host.
As a Repair Mechanism of DNA Double-Strand Breaks.
Although SINEs and LINEs and pseudogenes are abundant in eukaryotic genomes, indicating that reverse transcriptase-mediated phenomena are important in genome evolution, the mechanisms responsible for their spread are largely unknown. The results of two recent experiments with the yeast Saccharomyces cerevisiae (94, 95) have linked reverse transcriptase-mediated events with double-strand chromosome breaks in the absence of normal repair. This suggests a possible role for endogenous retroelements in the repair of double-strand chromosome breaks under certain circumstances. Note that, in this case, as in others described here, coadaptation may have grown out of apparently parasitic element behavior; double-strand chromosome breaks may simply represent an especially good target for efficient TE insertion, and in turn, these insertions may sometimes be the most efficient repair pathway available to the host. The net effect, rapid insertional repair of breaks, is expected to benefit both host and TE.
Mediation of the Concerted Evolution of Repetitive Gene Families.
Evidence for the ability of TEs to directly influence the constitution of repetitive DNA was provided by experiments using genetically marked P elements located in a subtelomeric repeat of D. melanogaster (96). After P element mobilization, the number of repeats frequently was observed to be altered, with decreases being more common than increases, due to unequal gene conversion events. Therefore, TEs may play an important role in the evolution of heterochromatin.
Changes in Genome Size.
As described above, TEs may represent a variable and sometimes surprisingly high proportion of genomes, particularly in plants. By means of variation in sheer bulk, it is possible that TEs affect variability in life history traits and related characteristics because of the correlation between genome size, cell size, and various aspects of plant life form, such as growth rate and developmental time (97).
Other Possible Functions.
The idea that heterochromatic clusters of nomadic elements are merely graveyards of dead transposons appears to be giving way to the idea that these regions may also be involved in a number of important cellular processes (25). These include developmentally programmed changes in DNA content, expression of heterochromatically embedded gene loci, and housekeeping functions such as chromosome pairing, sister chromatid adhesion, and centromere function. The TE content of these regions may be important for these processes in ways that are not yet understood.
Discussion
One of the most compelling questions that arise when considering the new data on the preponderance of TE-derived sequences in some plant genomes is how enormous numbers of TE copies can accumulate in a single genome. For example, how is it that a single TE can make up 10% of the maize genome? Obviously, the recombinogenic properties of TEs that are hypothesized to maintain a relatively low, constant copy number in other organisms are not relevant in these cases. It may be that, in the case of low copy elements transposing at relatively low frequencies, recombination is able to purge some elements from euchromatin, but the maize example suggests that there may be a vast number of elements interspersed between genes in many locations. Recombination, then, may not be a particularly effective mechanism for purging the vast majority of repetitive sequences, many of which are clearly not located in heterochromatin. It can be argued that, wherever there is a concentration of TE sequences, heterochromatin-like structural features begin to evolve to down-regulate their expression. In turn, this would also tend to reduce the frequency of removal by ectopic recombination.
These considerations motivate us to postulate that there are at least two types of elements that occupy two very different niches in the ecology of the genome: first, a type that preferentially inserts into regions distant from host gene sequences, such as heterochromatin or the regions between genes [e.g., the many retrotransposons found inserted between the genes on the third chromosome in maize (2)]; and second, a type that lives more dangerously by being more prone to insert into, or near, single copy sequences. We suggest that the first type escapes the “trap” of inactivation (via methylation or heterochromatinization) in regions outside of single copy host genes through the use of various buffer sequences; it has become specifically adapted to (or even makes up much of) these regions. As a strategy to minimize their potentially devastating effects on their hosts, these elements target regions in which recombination is minimal and where essential genes are scarce. The second type travels light and has evolved to take advantage of relatively accessible chromosomal architecture, a high concentration of transcription factors, host enhancer sequences, and horizontal transfer to maximize replication advantage. This type, represented by elements like Mu (which target single copy sequences) and P elements (at least 65% of insertions are located near enhancers) trades the disadvantage of an increased risk of negative selection for the advantages of occupying regions which are enriched for factors promoting efficient transcription and replication. This second type is postulated to be the one most likely to be discovered by geneticists (it is more likely to cause mutations) and also the one most likely to be lost through recombination (by targeting actively transcribed regions of the genome in which recombination is more frequent). We suggest that, when these elements insert in heterochromatin, they become inactive because they are not well adapted to that environment.
We therefore need to consider the possibility that there may be more than one strategy to being a transposon and that each strategy, although successful from an evolutionary perspective, has a very different dynamic. Each type would be expected to affect host evolution in a different way. Type 1 would affect the overall architecture of the host chromosome, rather than the specific expression characteristics of individual genes. In contrast, type 2 would participate more directly in changes in gene regulation, such as is observed at the Adh1 locus in maize.
A second area of considerable interest from an evolutionary perspective is the stress-induced mutability that is characteristic of some TEs (98). A gradualist argument leveled against the idea that regulatory changes resulting from TE-induced mutations may be important in evolution is that such “macromutations,” like Goldschmit’s hopeful monsters, would be unlikely to arise at the precise time when a new ecological niche became available (40). However, there is increasing evidence that TE-induced mutation rates are far from constant. High frequencies of mutations are expected to appear in waves, such as those resulting from hybrid dysgenesis that accompany element invasions of new populations or species. TE-induced mutations have been recorded to occur in transpositional bursts (99) whose cause is not well understood but is likely related to inbreeding and other forms of genomic or environmental stress, possibly akin to the genomic stress referred to by McClintock (100). For example, it appears that plant retroelements are normally quiescent but can be activated by stress (98), such as cell culturing (101) or microbial infection (102). We suggest that the proximal, or adaptive, function in these cases is to increase element copy number during periods of stress to ensure a high probability of transmission by those host variants that happen to survive. With respect to the evolution of the host, however, the preadaptive, or exaptive function is to provide variation during periods of stress. In this case, as in the other cases outlined above, the transposon does not have to “know” that it is contributing to the evolution of its host nor has it evolved to do so, but out of its elemental parasitic behavior arises the potential for both dramatic and subtle changes in the genome of its host.
Conclusions
We are only just beginning to glimpse the complexity of possible interactions in the coevolution of TEs and their hosts. A full understanding of the population and evolutionary dynamics of these interactions, and the consequences to hosts, must await the results of further research. However some tentative conclusions can be made on the basis of current information.
The primary parasitic nature of these sequences during their invasion of host populations is beyond dispute, but we believe that this does not by any means represent the whole story. A number of features of both TEs and their hosts can be interpreted as coadaptations to mitigate or abolish the reduction of fitness due to unbridled transposition. Furthermore, the number of well documented cases in which TE sequences have been coopted successfully by the host to provide a useful function is small but is growing rapidly. We suggest that the process by which elements and their hosts coevolve mutually beneficial strategies may lend itself to the production of genetic variation that would not otherwise have arisen.
Although the role of TEs in evolution may not turn out to be precisely what McClintock had in mind when she first described controlling elements in maize, the importance of their role in the evolution of gene regulation and other host functions may yet surprise us. To paraphrase Dobzhansky’s famous phrase, there is good reason to believe that “Nothing about mobile elements makes sense except in the light of evolution.”
Acknowledgments
We thank Dr. Zhijian Tu for comments on the manuscript. This work was supported by National Science Foundation Grant DEB9119349 to M.K. D.L. was supported by National Institutes of Health Training Program in Insect Science 1T32 AI07475.
ABBREVIATIONS
- TE
transposable elements
- MITE
miniature inverted-repeat TE
References
- 1.McClintock B. Carnegie Inst Wash Yearbook. 1948;47:155–169. [Google Scholar]
- 2.SanMiguel P, Tikhonov A, Jin Y K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, Bennetzen J L. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 3.Pimpinelli S, Berloco M, Fanti L, Dimitri P, Bonaccorsi S, Marchetti E, Caizzi R, Caggesse C, Gatti M. Proc Natl Acad Sci USA. 1995;92:3804–3808. doi: 10.1073/pnas.92.9.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finnegan D J. Curr Opin Genet Dev. 1992;2:861–867. doi: 10.1016/s0959-437x(05)80108-x. [DOI] [PubMed] [Google Scholar]
- 5.McClure M A. Reverse Transcriptase. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 425–443. [Google Scholar]
- 6.Bucheton A. Trends Genet. 1995;11:349–353. doi: 10.1016/s0168-9525(00)89105-2. [DOI] [PubMed] [Google Scholar]
- 7.Wessler S, Bureau T E, White S E. Curr Opin Genet Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- 8.Unsal K, Morgan G T. J Mol Biol. 1995;248:812–823. doi: 10.1006/jmbi.1995.0262. [DOI] [PubMed] [Google Scholar]
- 9.Morgan J T. J Mol Biol. 1995;254:1–5. doi: 10.1006/jmbi.1995.0593. [DOI] [PubMed] [Google Scholar]
- 10.Smit A F A, Riggs A D. Proc Natl Acad Sci USA. 1996;93:1443–1448. doi: 10.1073/pnas.93.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tu, J. (1997) Proc. Natl. Acad. Sci. USA, in press.
- 12.Britten R J. Proc Natl Acad Sci USA. 1996;93:9374–9377. doi: 10.1073/pnas.93.18.9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engels W R. In: Transposable Elements. Saedler H, Gierl A, editors. Berlin: Springer; 1996. pp. 103–123. [Google Scholar]
- 14.Bregliano J C, Kidwell M G. In: Mobile Genetic Elements. Shapiro J A, editor. New York: Academic; 1983. pp. 363–410. [Google Scholar]
- 15.Rubin G M, Kidwell M G, Bingham P M. Cell. 1982;29:987–994. doi: 10.1016/0092-8674(82)90462-7. [DOI] [PubMed] [Google Scholar]
- 16.McClintock B. Cold Spring Harbor Symp Quant Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Wessler S, Baran G, Varagona M, Dellaporta S. EMBO J. 1986;5:2427–2432. doi: 10.1002/j.1460-2075.1986.tb04517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wessler S, Baran G, Varagona M. Science. 1987;237:916–918. doi: 10.1126/science.3039661. [DOI] [PubMed] [Google Scholar]
- 19.Corces V G, Geyer P K. Trends Genet. 1991;7:86–90. doi: 10.1016/0168-9525(91)90277-W. [DOI] [PubMed] [Google Scholar]
- 20.Lister C, Jackson D, Martin C. Plant Cell. 1993;5:1541–1553. doi: 10.1105/tpc.5.11.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kloeckener-Gruissem B, Vogel J M, Freeling M. EMBO J. 1992;11:157–166. doi: 10.1002/j.1460-2075.1992.tb05038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kloeckener-Gruissem B, Freeling M. Proc Natl Acad Sci USA. 1995;92:1836–1840. doi: 10.1073/pnas.92.6.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greene B, Walko R, Hake S. Genetics. 1994;138:1275–1285. doi: 10.1093/genetics/138.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bradley D, Carpenter R, Sommer H, Hartley N, Coen E. Cell. 1993;72:85–95. doi: 10.1016/0092-8674(93)90052-r. [DOI] [PubMed] [Google Scholar]
- 25.Arkhipova I R, Lyubomirskaya N V, Ilyin Y V. Drosophila Retrotransposons. Austin, TX: Landes; 1995. [Google Scholar]
- 26.Charlesworth B, Langley C H. Annu Rev Genet. 1989;23:251–287. doi: 10.1146/annurev.ge.23.120189.001343. [DOI] [PubMed] [Google Scholar]
- 27.Vaury C, Bucheton A, Pelisson A. Chromosoma. 1989;98:215–224. doi: 10.1007/BF00329686. [DOI] [PubMed] [Google Scholar]
- 28.Tchurikov N A, Zelentsova E S, Georgiev G P. Nucleic Acids Res. 1980;8:1243–1258. doi: 10.1093/nar/8.6.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tchurikov N A, Ilyin Y V, Skryabin K G, Anan’ev E V, Bayev A A, Krayev A S, Zelentsova E S, Kulguskin V V, Lyubomirskaya N V, Georgiev G P. Cold Spring Harbor Symp Quant Biol. 1981;45:655–665. doi: 10.1101/sqb.1981.045.01.083. [DOI] [PubMed] [Google Scholar]
- 30.Banga S S, Velazquez A, Boyd J B. Mutat Res. 1991;255:79–88. doi: 10.1016/0921-8777(91)90020-p. [DOI] [PubMed] [Google Scholar]
- 31.Charlesworth B, Sniegowski P, Stephan W. Nature (London) 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- 32.Lyttle T W, Haymer D S. In: Transposable Elements and Evolution. McDonald J F, editor. Dordrecht, The Netherlands: Kluwer; 1993. [Google Scholar]
- 33.Simmons G M. Mol Biol Evol. 1992;9:1050–1060. doi: 10.1093/oxfordjournals.molbev.a040774. [DOI] [PubMed] [Google Scholar]
- 34.Mackay T F C. Genet Res. 1987;49:225–233. [Google Scholar]
- 35.Mackay T F, Lyman R F, Jackson M S. Genetics. 1992;130:315–332. doi: 10.1093/genetics/130.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torkamanzehi A, Moran C, Nicholas F W. Genetics. 1992;131:73–78. doi: 10.1093/genetics/131.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark A G, Wang L, Hulleberg T. Genetics. 1995;139:337–348. doi: 10.1093/genetics/139.1.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Britten R J. Mol Phylogenet Evol. 1996;5:13–17. doi: 10.1006/mpev.1996.0003. [DOI] [PubMed] [Google Scholar]
- 39.Britten, R. J. (1997) Gene, in press.
- 40.McDonald J F. Trends Ecol Evol. 1995;10:123–126. doi: 10.1016/s0169-5347(00)89012-6. [DOI] [PubMed] [Google Scholar]
- 41.Doolittle W F, Sapienza C. Nature (London) 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 42.Orgel, L. E. & Crick, F. H. C. (1980) Nature (London) 284. [DOI] [PubMed]
- 43.Hickey D A. Genetics. 1982;101:519–531. doi: 10.1093/genetics/101.3-4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plasterk R A. In: Mobile Genetic Elements. Sherratt D J, editor. Oxford: IRL; 1995. pp. 18–37. [Google Scholar]
- 45.Kiyasu P K, Kidwell M G. Genet Res. 1984;44:251–259. doi: 10.1017/s0016672300026495. [DOI] [PubMed] [Google Scholar]
- 46.Kidwell M G. Ann Rev Genet. 1993;27:235–256. doi: 10.1146/annurev.ge.27.120193.001315. [DOI] [PubMed] [Google Scholar]
- 47.Miller W J, Kruckenhauser L, Pinsker W. In: Transgenic Organisms: Biological and Social Implications. Tomiuk J, Woehrmann K, Sentker A, editors. Basel: Birkhaeuser; 1996. pp. 21–35. [Google Scholar]
- 48.Hurst G D D, Hurst L D, Majerus M E N. Nature (London) 1992;356:659–660. doi: 10.1038/356659a0. [DOI] [PubMed] [Google Scholar]
- 49.Kidwell M G. J Hered. 1994;85:339–346. doi: 10.1093/oxfordjournals.jhered.a111478. [DOI] [PubMed] [Google Scholar]
- 50.Robertson H M. Nature (London) 1993;362:241–245. doi: 10.1038/362241a0. [DOI] [PubMed] [Google Scholar]
- 51.Lohe A R, Moriyama E N, Lidholm D A, Hartl D L. Mol Biol Evol. 1995;12:62–72. doi: 10.1093/oxfordjournals.molbev.a040191. [DOI] [PubMed] [Google Scholar]
- 52.Clark J B, Maddison W P, Kidwell M G. Mol Biol Evol. 1994;11:40–50. doi: 10.1093/oxfordjournals.molbev.a040091. [DOI] [PubMed] [Google Scholar]
- 53.Spradling A C, Stern D M, Kiss I, Roote J, Laverty T, Rubin G M. Proc Natl Acad Sci USA. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bellen H, O’Kane C J, Wilson C, Grossniklaus U, Pearson R K, Gehring W J. Genes Dev. 1989;3:1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- 55.Ji H, Moore D P, Blomberg M A, Braiterman L T, Voytas D F, Natsoulis G, Boeke J D. Cell. 1993;73:1007–1018. doi: 10.1016/0092-8674(93)90278-x. [DOI] [PubMed] [Google Scholar]
- 56.Hallick R B, Hong L, Drager R G, Favreau M R, Montfort A, Orsat B, Spielmann A, Stutz E. Nucleic Acids Res. 1993;21:3537–3544. doi: 10.1093/nar/21.15.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fridell R A, Pret A M, Searles L L. Genes Dev. 1990;4:559–566. doi: 10.1101/gad.4.4.559. [DOI] [PubMed] [Google Scholar]
- 58.Purugganan M, Wessler S. Genetica. 1992;86:295–303. doi: 10.1007/BF00133728. [DOI] [PubMed] [Google Scholar]
- 59.Rushforth A M, Anderson P. Mol Cell Biol. 1996;16:422–429. doi: 10.1128/mcb.16.1.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laski F A, Rio D C, Rubin G M. Cell. 1986;44:7–19. doi: 10.1016/0092-8674(86)90480-0. [DOI] [PubMed] [Google Scholar]
- 61.Tseng J C, Zollman S, Chain A C, Laski F A. Mech Dev. 1991;35:65–72. doi: 10.1016/0925-4773(91)90042-5. [DOI] [PubMed] [Google Scholar]
- 62.Bingham P M, Chou T B, I, M, Zachar Z. Trends Genet. 1988;4:134–138. doi: 10.1016/0168-9525(88)90136-9. [DOI] [PubMed] [Google Scholar]
- 63.Chomet P S, Wessler S, Dellaporta S L. EMBO J. 1987;6:295–302. doi: 10.1002/j.1460-2075.1987.tb04753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chandler V, Walbot V. Proc Natl Acad Sci USA. 1986;83:1767–1771. doi: 10.1073/pnas.83.6.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bennetzen J L. In: Transposable Elements. Saedler H, Gierl A, editors. Berlin: Springer; 1996. pp. 195–229. [Google Scholar]
- 66.Charlesworth B, Langley C H. Genetics. 1986;112:359–383. doi: 10.1093/genetics/112.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brookfield J F Y. In: Mobile Genetic Elements. Sherratt D J, editor. Oxford: IRL; 1995. pp. 130–153. [Google Scholar]
- 68.Robertson H M, Engels W R. Genetics. 1989;123:815–824. doi: 10.1093/genetics/123.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Misra S, Rio D C. Cell. 1990;62:269–284. doi: 10.1016/0092-8674(90)90365-l. [DOI] [PubMed] [Google Scholar]
- 70.Black D M, Jackson M S, Kidwell M G, Dover G A. EMBO J. 1987;6:4125–4135. doi: 10.1002/j.1460-2075.1987.tb02758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jackson M S, Black D M, Dover G A. Genetics. 1988;120:1003–1013. doi: 10.1093/genetics/120.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rasmusson K E, Raymond J D, Simmons M J. Genetics. 1993;133:605–622. doi: 10.1093/genetics/133.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fedoroff N, Schlappi M, Raina R. Bioessays. 1995;17:291–297. doi: 10.1002/bies.950170405. [DOI] [PubMed] [Google Scholar]
- 74.Schlappi M, Raina R, Fedoroff N. Cell. 1994;77:427–437. doi: 10.1016/0092-8674(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 75.Cuypers H, Dash S, Peterson P A, Saedler H, Gierl A. EMBO J. 1988;7:2953–2960. doi: 10.1002/j.1460-2075.1988.tb03157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McClintock B. Carnegie Inst Wash Yearbook. 1958;57:415–429. [Google Scholar]
- 77.Gdula D A, Gerasimova T I, Corces V G. Proc Natl Acad Sci USA. 1996;93:9378–9383. doi: 10.1073/pnas.93.18.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Csink A, McDonald J F. Genetics. 1990;126:375–385. doi: 10.1093/genetics/126.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.King M C, Wilson A C. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 80.Britten R J, Davidson E H. Q Rev Biol. 1971;46:111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- 81.Li X, Noll M. Nature (London) 1994;367:83–87. doi: 10.1038/367083a0. [DOI] [PubMed] [Google Scholar]
- 82.Fincham J R S, Sastry G R K. Annu Rev Genet. 1974;8:15–50. doi: 10.1146/annurev.ge.08.120174.000311. [DOI] [PubMed] [Google Scholar]
- 83.Bureau T E, Ronald P C, Wessler S R. Proc Natl Acad Sci USA. 1996;93:8524–8529. doi: 10.1073/pnas.93.16.8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pelisson A, Song S U, Prud’homme N, Smith P A, Bucheton A, Corces V G. EMBO J. 1994;13:4401–4411. doi: 10.1002/j.1460-2075.1994.tb06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ding D, Lipshitz H D. Genet Res. 1994;64:167–181. doi: 10.1017/s0016672300032833. [DOI] [PubMed] [Google Scholar]
- 86.Hambor J E, Mennone J, Coon M E, Hanke J H, Kavathas P. Mol Cell Biol. 1993;13:7056–7070. doi: 10.1128/mcb.13.11.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stavenhagen J B, Robins D M. Cell. 1988;55:247–254. doi: 10.1016/0092-8674(88)90047-5. [DOI] [PubMed] [Google Scholar]
- 88.Anderson R, Britten R J, Davidson E H. Dev Biol. 1994;163:11–18. doi: 10.1006/dbio.1994.1119. [DOI] [PubMed] [Google Scholar]
- 89.Miller, W. J., McDonald, J. F. & Pinsker, W. (1997) Genetica, in press. [PubMed]
- 90.Miller W J, Paricio N, Hagemann S, Martinez-Sebastian M J, Pinsker W, de Frutos R. Gene. 1995;156:167–174. doi: 10.1016/0378-1119(95)00013-v. [DOI] [PubMed] [Google Scholar]
- 91.Hong L, Hallick R B. Genes Dev. 1994;8:1589–1599. doi: 10.1101/gad.8.13.1589. [DOI] [PubMed] [Google Scholar]
- 92.Biessmann H, Valgeirsdottir K, Lofsky A, Chin C, Ginther B, Levis R W, Pardue M L. Mol Cell Biol. 1992;12:3910–3918. doi: 10.1128/mcb.12.9.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pardue M L, Danilevskaya O N, Lowenhaupt K, Slot F, Traverse K L. Trends Genet. 1996;12:48–52. doi: 10.1016/0168-9525(96)81399-0. [DOI] [PubMed] [Google Scholar]
- 94.Moore J K, Haber J E. Nature (London) 1996;383:644–646. doi: 10.1038/383644a0. [DOI] [PubMed] [Google Scholar]
- 95.Teng S C, Kim B, Gabriel A. Nature (London) 1996;383:641–644. doi: 10.1038/383641a0. [DOI] [PubMed] [Google Scholar]
- 96.Thompson-Stewart D, Karpen G H, Spradling A C. Proc Natl Acad Sci USA. 1994;91:9042–9046. doi: 10.1073/pnas.91.19.9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smyth D R. In: Control of Plant Gene Expression. Verma D P S, editor. Boca Raton, FL: CRC; 1993. [Google Scholar]
- 98.Wessler S R. Curr Biol. 1996;6:959–961. doi: 10.1016/s0960-9822(02)00638-3. [DOI] [PubMed] [Google Scholar]
- 99.Gerasimova T I, Matjunima L V, Mizrokhi L J, Georgiev G P. EMBO J. 1985;4:3773–3779. doi: 10.1002/j.1460-2075.1985.tb04147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McClintock B. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 101.Pouteau S, Huttner E, Grandbastien M A, Caboche M. EMBO J. 1991;10:1911–1918. doi: 10.1002/j.1460-2075.1991.tb07717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pouteau S, Grandbastien M A, Boccara M. Plant J. 1994;5:535–542. [Google Scholar]