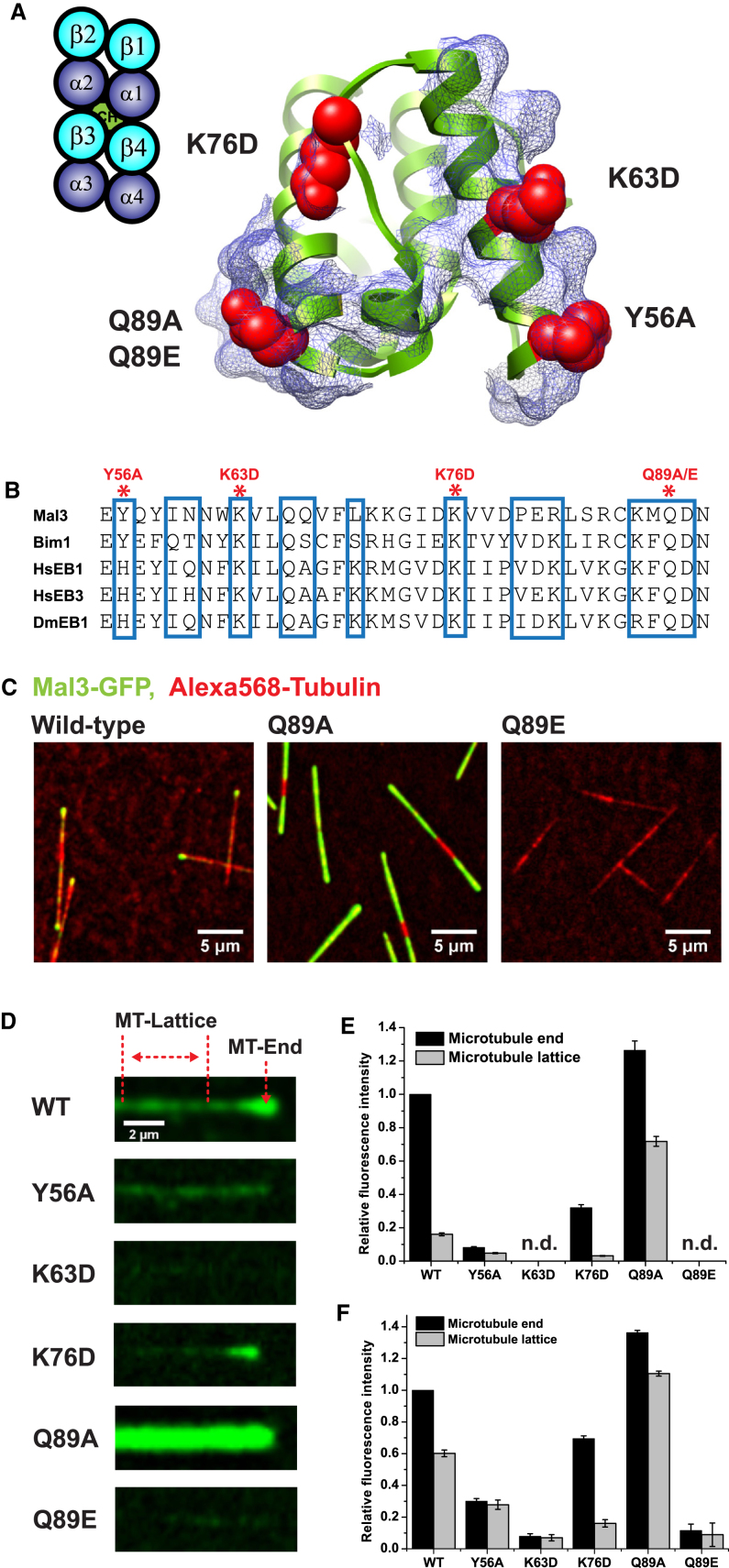
Figure 5.
Validation of EB-Tubulin Contact Sites using Reconstituted Dynamic Microtubule-End Tracking
(A) Localization of single mutated residues on the Mal3 microtubule-binding surface: residues chosen for mutations (red) in the context of all Mal3 residues within 5 Å of tubulin (rendered as a blue mesh).
(B) Extract of a sequence alignment of different EBs: Mal3 residues contacting the microtubule (blue boxes) and residues that were mutated in this study (red asterisks).
(C) TIRF microscopy images of Alexa 568-labeled microtubules (red) grown in the presence of GTP and wild-type (WT) or mutant Mal3-GFP (green) as indicated.
(D) TIRF microscopy images illustrating the differences between the Mal3-GFP fluorescence signal (green) for WT and the different mutants on single microtubules. Imaging conditions and display settings are identical for all six experiments. The regions used to measure Mal3-GFP intensity at the microtubule end and on the lattice are indicated by red lines.
(E and F) Quantification of Mal3-GFP intensities at the microtubule ends and on the lattice. Error bars represent standard error of the mean (SEM). (E) Mean intensities relative to the mean WT intensity at growing microtubule ends are shown. Concentrations were 30 nM Mal3-GFP, 22 μM tubulin, and 1 mM GTP. (F) Same as (E) with 300 nM Mal3-GFP.