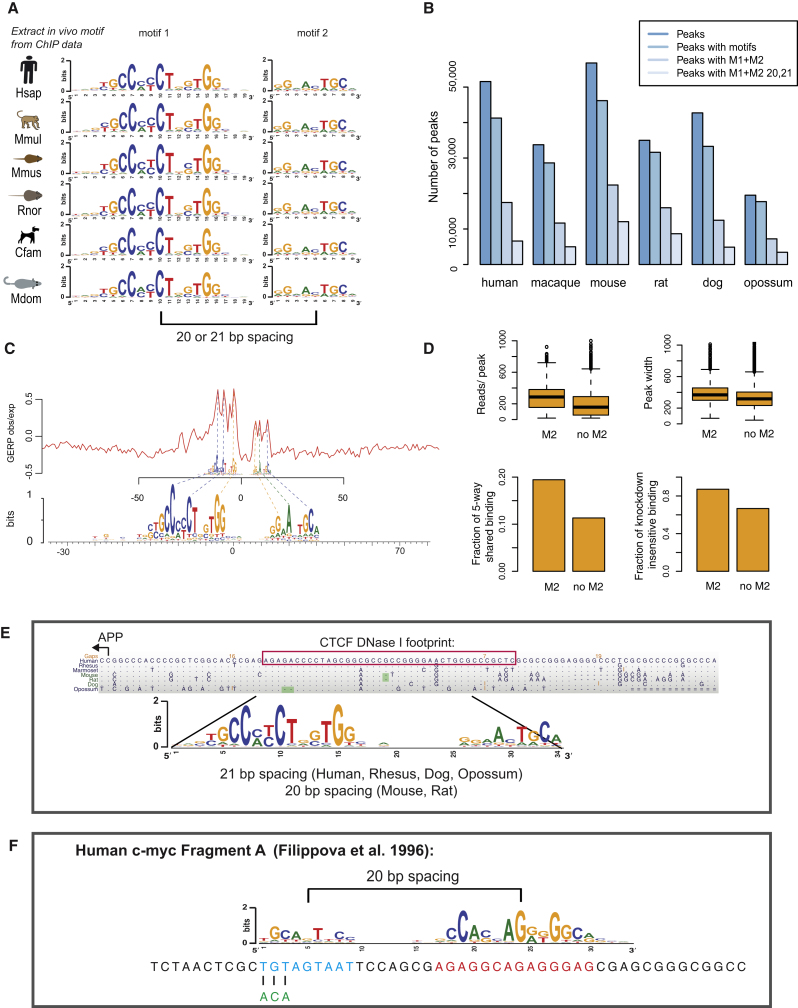
Figure 2.
CTCF Binding Often Occurs at a Highly Conserved Motif, Consisting of a Two-Part Profile
(A) Motifs (M1 and M2) identified de novo from CTCF-binding events.
(B) Binding event counts and number of binding events with at least one motif (M1 and M1+M2) in all six species. M1+M2 20,21 represents the preferred spacing patterns of these two submotifs.
(C) The DNA sequence constraint around the CTCF motif in human was plotted by observed/expected genomic evolutionary rate profiling (red, GERP) scores (Cooper et al., 2005). The frequencies of unchanged bases in five-way shared CTCF-binding events are shown as position weight matrix (PWM) below the GERP profile.
(D) Peaks containing the M2 motif in preferred spacing are stronger in ChIP enrichment both by read count and peak width, are more highly shared among mammals, and are resistant to RNAi-mediated knockdown.
(E) A multiple mammalian sequence alignment of a CTCF peak at the APP gene is shown. The DNase I footprint (red box, Quitschke et al., 2000) encompasses a complete 34 bp M1 and M2 CTCF motif.
(F) DNA sequence of the human c-myc promoter (Human c-myc Fragment A) bound by CTCF in vivo and in vitro (Filippova et al., 1996). The sequence contains the canonical M1 CTCF motif (red) and the M2 motif (blue). A 3 bp mutation in the M2 motif that eliminates CTCF binding in vitro is indicated in green.
See also Figure S2.