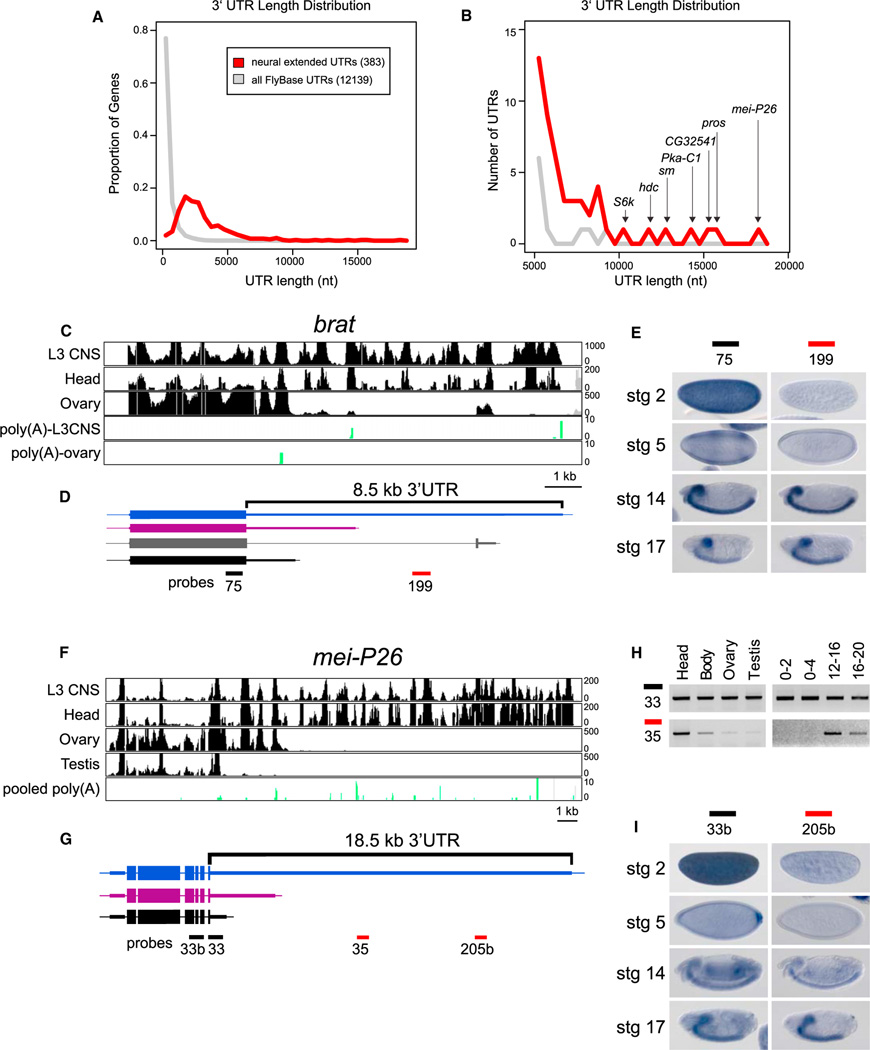
Figure 3. Unusually Long 3′ UTR Extensions in Neural Genes.
(A) Distribution of 3′ UTR lengths of 383 transcripts that exhibit neural lengthening, relative to the longest annotated 3′ UTRs of all FlyBase 5.32 genes.
(B) Focusing on the tail of the distribution, the neural 3′ UTR extensions comprise most of the longest 3′ UTRs in the Drosophila transcriptome.
(C and D) RNA-seq tracks from indicated tissues (C) and transcript models for brat (D). A splice variant that alters the last few amino acids of Brat was apparent from RNA-seq data (gray), but this isoform was not differentially expressed.
(E) In situ hybridization revealed that both proximal and distal probes detect CNS expression in germband retracted embryos and late stage embryos, but that the proximal probe alone detects maternal deposition and blastoderm expression in two stripes along the anterior-posterior axis.
(F and G) RNA-seq tracks from indicated tissues (F) and gene models for mei-P26 (G).
(H) Semiquantitative RT-PCR indicates that RNA corresponding to the 3′ end of the coding sequence and proximal 3′ UTR can be readily detected in all tissues and embryonic time-points examined, whereas an amplicon ~8 kb into the 3′ UTR was predominantly detected in heads and late stage embryos.
(I) In situ hybridization reveals that both proximal and distal probes detect late stage expression in the brain and ventral nerve cord, but that the proximal probe alone detects maternal deposition at stage 2, posterior expression at stage 5, and ubiquitous expression at stage 14.