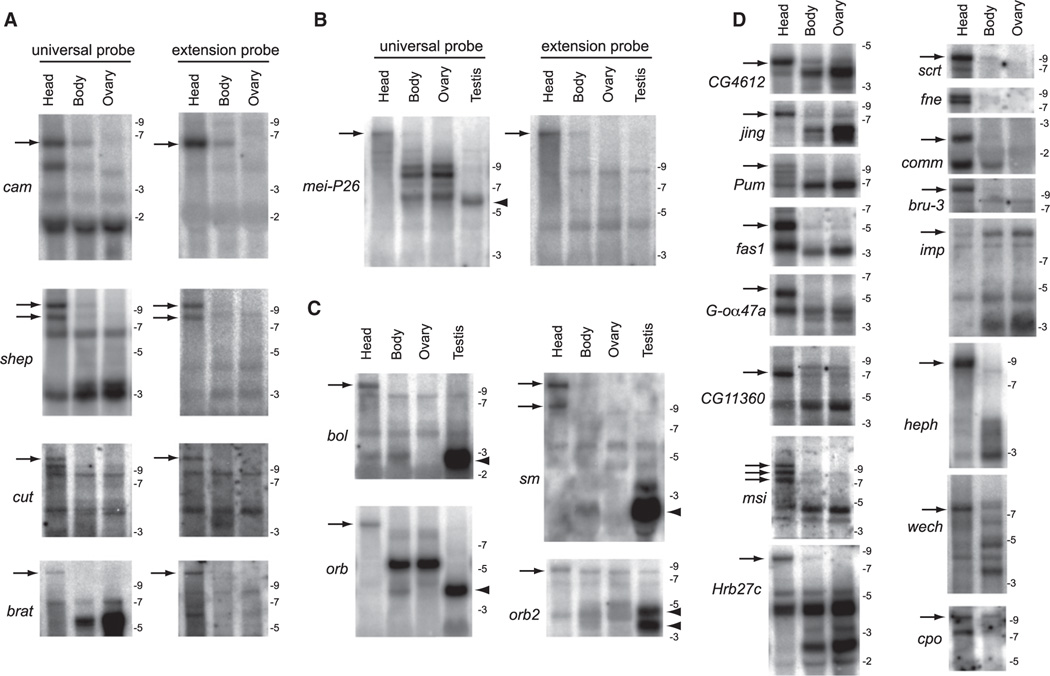
Figure 4. Northern Analysis Validates Stable Transcripts Corresponding to Strongly Distal APA Isoforms in Drosophila Heads.
(A) Comparison of proximal “universal” probes and distal “extension” probes for a selection of exceptionally long inferred APA isoforms. In each case, a similarly sized band(s) is detected only in head samples with the paired probes (arrows), demonstrating that they connect a common 3′ UTR-extended transcript. In many cases, the sizes of these discrete lengthened transcripts far exceed the largest molecular weight marker on the RNA ladder (9 kb).
(B) Comparison of proximal universal probes and distal extension probes for mei-P26. Note the 3′ UTR lengthening in head and 3′ UTR shortening in testis relative to body/ovary samples.
(C) Examples of APA transcripts exhibiting neural extension (arrows) and testis shortening (arrowheads), often with intermediate-sized transcripts in body and/or ovary.
(D) A broad selection of other transcripts that exhibit 3′ UTR-extended isoforms predominantly or exclusively in head (arrows), with the exception of imp. For bands that were outside the range of the RNA ladder, the estimated size of the longest 3′ UTR isoform in head was calculated based on RNA-seq coverage from head library and existing RefSeq mRNA annotations: shep, 9.4 kb; cut, 11.6 kb; brat, 11.7 kb; mei-P26, 22.9kb; bol, 10.9 kb; orb, 8.9 kb; sm, 12.7 kb; pum, 10.1 kb; msi, 10.9 kb; scrt, 9.1 kb; wech, 7.6 kb; imp, 11.8 kb; cpo, 9.3 kb; bru-3, 9.6 kb.