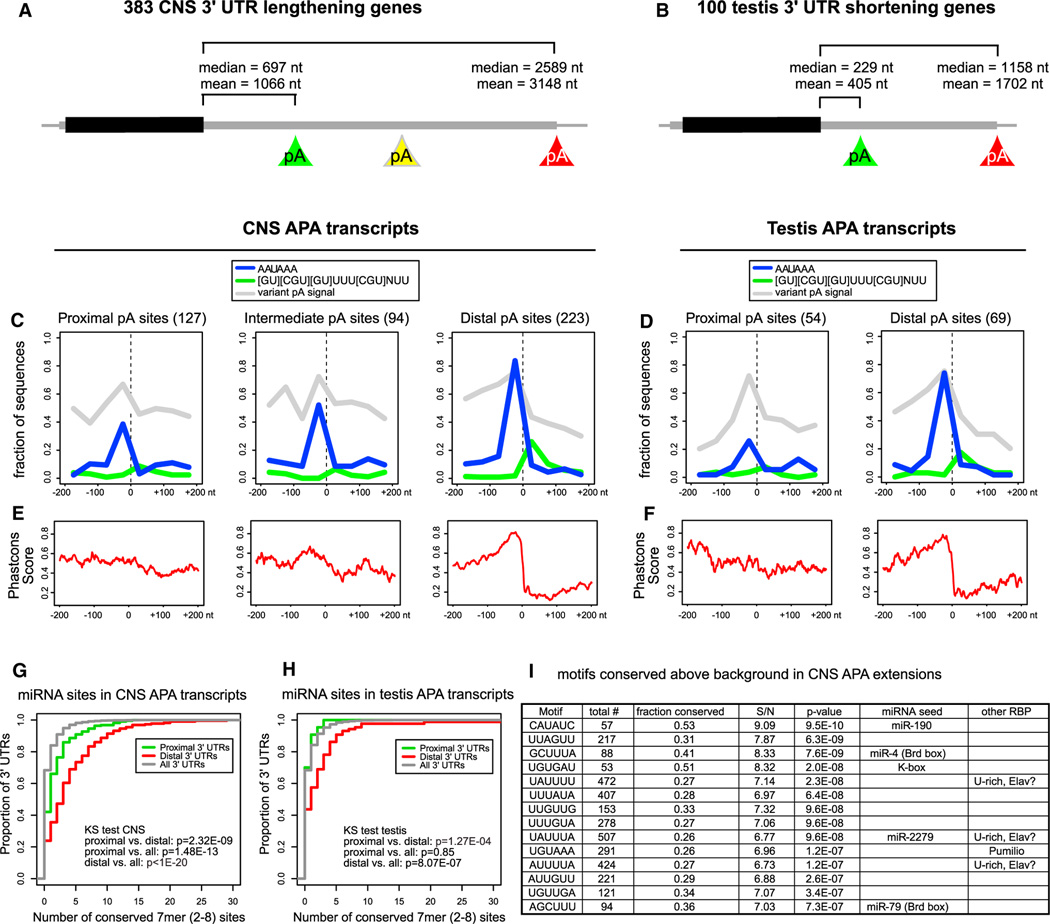
Figure 6. Quality and Conservation of Alternative Poly(A) Sites and miRNA Sites.
(A and B) Average 3′ UTR lengths of the proximal and distal transcript isoforms for genes exhibiting CNS 3′ UTR lengthening (A) and testis 3′ UTR shortening (B).
(C and D) Sequence motifs involved in polyadenylation around poly(A) sites identified by poly(A)-spanning reads: Canonical PAS (in blue), GU-rich DSE (in green) and variant PAS (in gray). (C) For CNS extended genes, distal sites are enriched for the canonical PAS compared to proximal sites (Fisher’s exact test p value = 4.9E-16), whereas the frequency at the intermediate poly(A) sites lies in between. The distal poly(A) sites are also enriched for the DSE motif, compared to the proximal and intermediate poly(A) sites (Fisher’s exact test p value = 7.2E-4). All poly(A) sites show similar collective levels of variant PAS; see also Figure S5 for analysis of individual PAS variants. (D) Testis APA transcripts display similar patterns of motif occurrence and conservation as the CNS transcripts (enrichment of AAUAAA in distal versus proximal sites, Fisher’s exact test p value = 2.6E-4).
(E) PhastCons conservation scores in the vicinity of poly(A) sites. Proximal and intermediate CNS poly(A) sites have intermediate conservation levels whereas distal sites have a region of high conservation just upstream of the poly(A) site and a region of low conservation downstream of the poly(A) site.
(F) Proximal poly(A) sites in testis transcripts have intermediate conservation levels whereas the distal sites have a region of high conservation just upstream of the poly(A) site and region of low conservation downstream of the poly(A) site.
(G) Cumulative distribution of numbers of miRNA target sites across all Drosophila 3′ UTRs, compared with the proximal and distal APA 3′ UTR variants of the 383 transcripts exhibiting neural lengthening. The distal extensions bear significantly more miRNA binding sites than Drosophila 3′ UTRs do in general.
(H) Cumulative distribution of miRNA target site numbers in the 100 testis APA transcripts.
(I) Motifs conserved above background in 3′ UTR extensions of neural transcripts. Shown are the top 6-mer motifs conserved above background in greater than or equal to seven Drosophilid species among the 383 3′ UTR extensions of neural transcripts. Many sites correspond to miRNA seeds or neural RBP sites (e.g., Pumilio and U-rich sequences that may potentially include Elav binding sites). A complete list of 6-mer and 7-mer motifs conserved above background are shown in Table S3. S/N, signal/noise ratio.