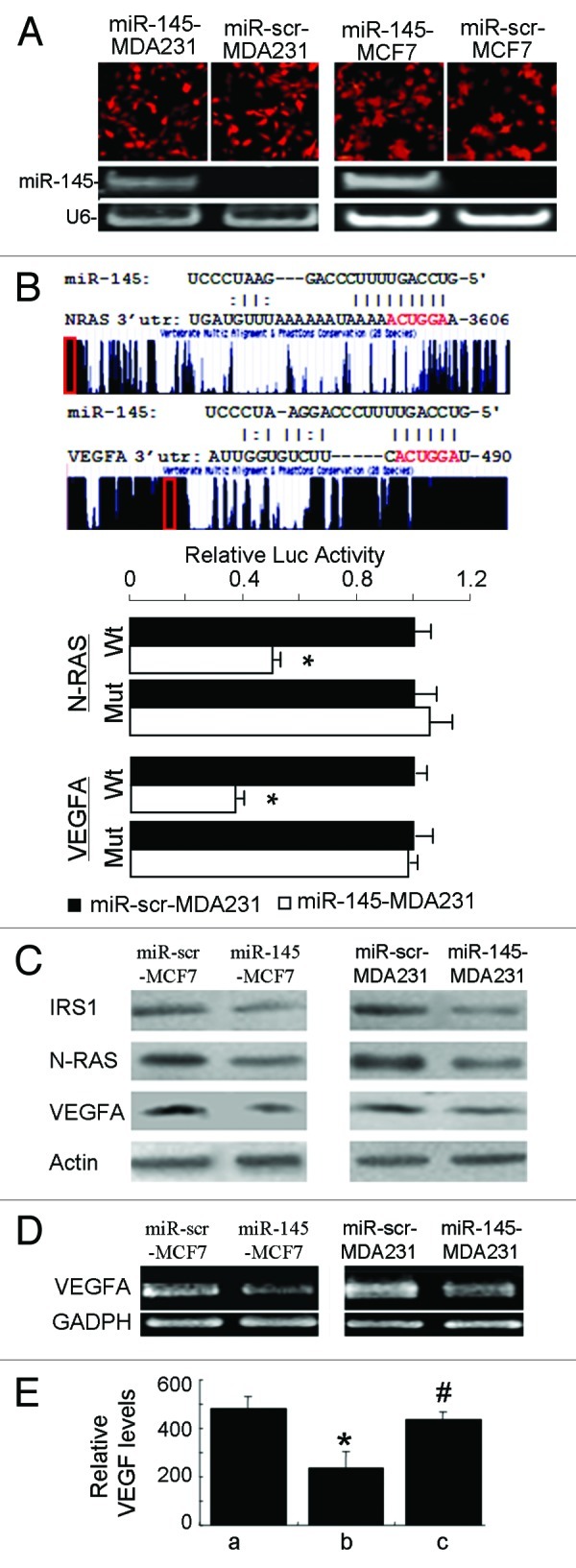
Figure 3. miR-145 targets N-RAS and VEGF-A. (A) Fluorescent images showed high transduction efficiency of lentivirus carrying miR-145 or miR-scr (upper panel). The miR-145 expression levels were detected by semi-quantitative RT-PCR (lower panel). (B) Schematic diagrams of putative miR-145 binding sites in the 3′UTRs of N-RAS and VEGF-A were shown in upper panel. The possible miR-145 binding sites in the 3′UTRs of the target genes were searched using algorithm KeyTar miRNA target prediction algorithm. The mutated nucleotides are shown in red. Luciferase activities of N-RAS and VEGFA reporter constructs in MDA-MB-231 cells transfected with miR-145 or miR-scr were normalized to cotransfected renilla luciferase plasmid activity (lower panel). *p < 0.05 (n = 3). Wt, miR-145 binding site wild type; Mut, miR-145 binding site mutant. (C) Immunoblotting analysis of IRS1, N-RAS and VEGF-A levels in lenti-miR-145 infected cells. (D) Semi-quantitative RT-PCR analysis for VEGF-A expression showed that it was significantly suppressed in lenti-miR-145- transduced MCF7 and MDA231 cells. GAPDH served as an internal control. (E) Forced expression of N-RAS reversed miR-145-inhibited VEGF expression. Relative VEGF levels were measured using ELISA in MCF7 cells infected by lenti-scr (a), miR-145 (b) and miR-145 with N-RAS plasmid (c) 48 h after transfection. * p < 0.01 (n = 3), b vs. a; #p < 0.01 (n = 3), c vs. b.
