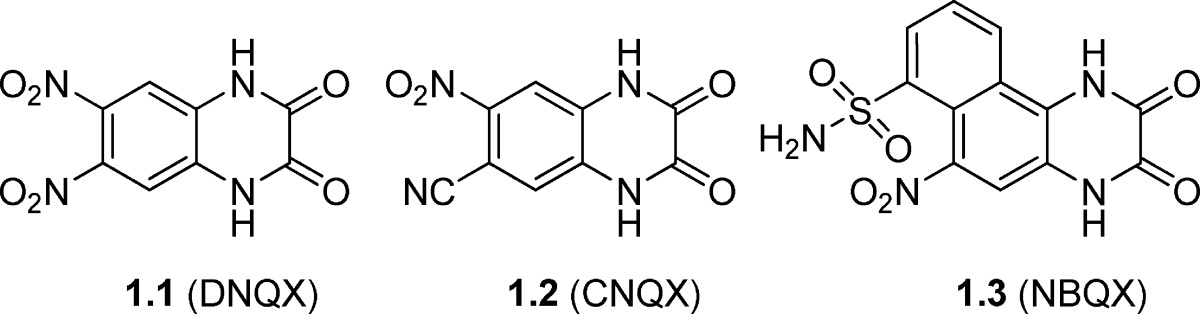
Table 1. Chemical Structures and Pharmacological Profile of Quinoxaline-2,3-dione Analogues 1.1−1.3a.
| AMPA | KA | NMDA | GluK1 |
GluK2 |
GluK3 | GluK2/5 |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| cmpd | func.b | func.b | func.b | bind.c | func.d | bind.e | func.d | bind.c | bind.c | func.d |
| 1.1 (DNQX) | 0.25f | 0.53f | 4.1f | −h | −h | 0.35 | −h | −h | −h | −h |
| 1.2 (CNQX) | 0.40f | 0.27f | 13f | −h | 8.0 | 0.53 | 18 | −h | −h | 72 |
| 1.3 (NBQX) | 0.063g | 0.078g | >300g | 12 | 25 | 0.87 | 21 | 24 | 0.6 | 87 |
Data for KA receptor subunits were obtained in the presence of ConA. All values in [μM].
Ki values for inhibition of currents activated by kainate (100 μM), AMPA (30 μM), or NMDA (30 μM/glycine (3 μM)) in Xenopus oocytes injected with mRNA from rat cortex.
Ki values for displacement of [3H]KA radioligand at human KA receptors expressed in HEK293 cells; from ref (56).
IC50 values for inhibition of 100 μM glutamate-induced Ca2+ influx in HEK293 cells expressing human KA receptors; from ref (50).
Ki values for displacement of [3H]KA radioligand at homomeric receptors in BHK cells; from ref (51).
From ref (57).
From ref (58).
Not tested.