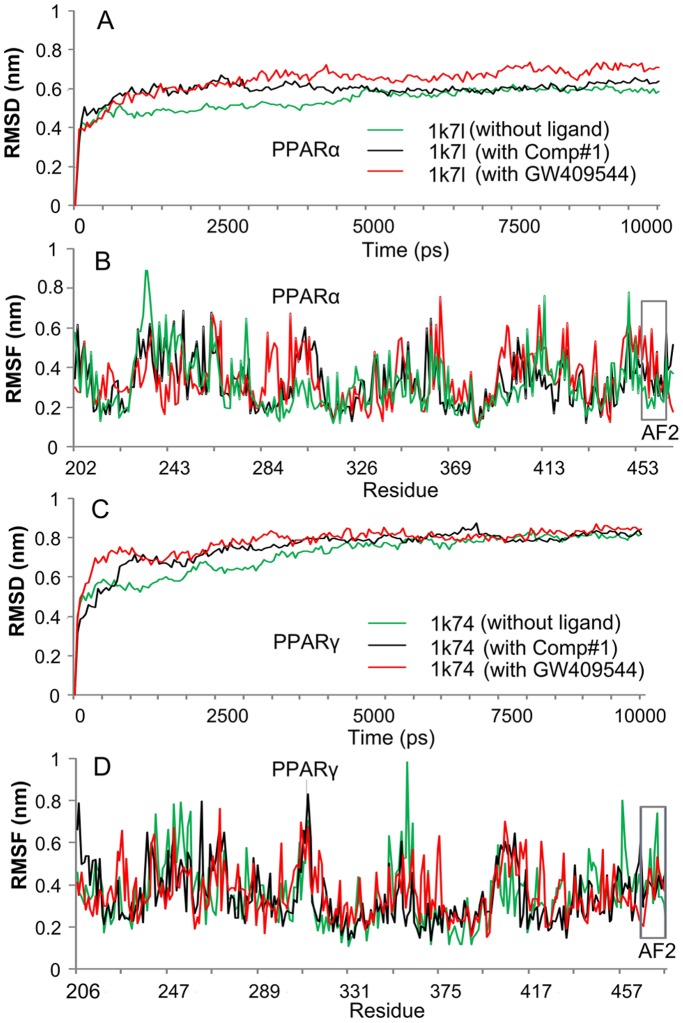
Figure 3. Illustration to show the outcomes of molecular dynamics simulations for Comp#1 ranked number 1 in Table 1 .
(A) The RMSD (root mean square deviation) of all backbone atoms for the receptor PPARα. (B) The RMSF (root mean square fluctuation) of the side-chain atoms for the receptor PPARα. (C) The RMSD (root mean square deviation) of all backbone atoms for the receptor PPARγ. (D) The RMSF (root mean square fluctuation) of the side-chain atoms for the receptor PPARγ. The green line indicates the outcome for the system of the receptor alone without any ligand, the red line for that of the receptor with the ligand GW409544, and the black line for that of the receptor with the ligand Comp#1. The curves involved with the AF2 helix region are framed with grey line. See the text for further explanation.