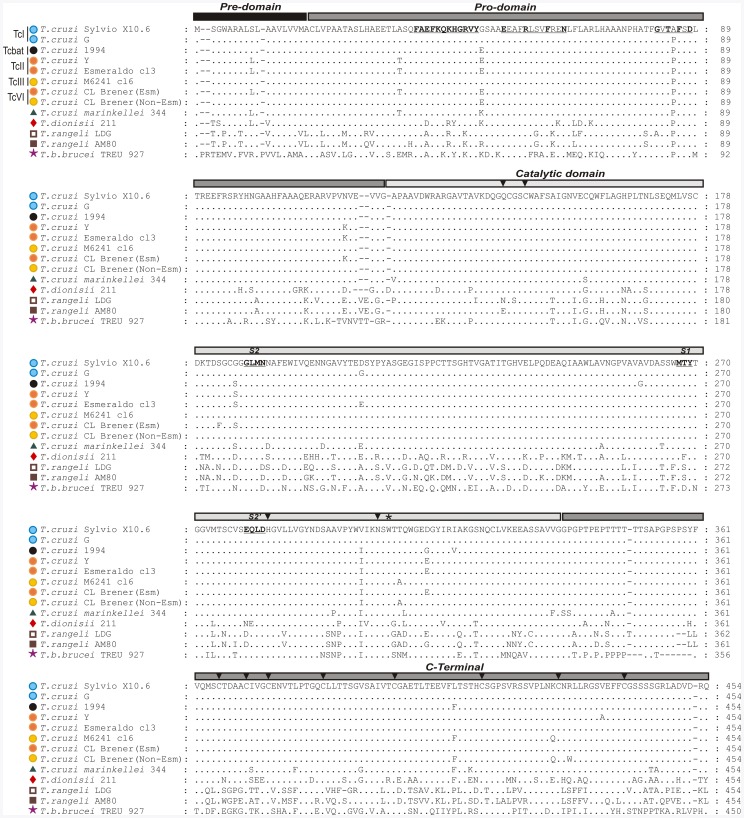
Figure 1. Alignment of predicted amino acid sequences from entire cruzipain of T. cruzi (TcI, TcII, TcIII, TcVI and Tcbat) and homologues from T. cruzi-like (T. c. marinkellei and T. dionisii), T. rangeli and T. b. brucei.
Pre, pro, catalytic domain and C-terminal extension amino acid sequences of cruzipain genes from T. cruzi Sylvio X10.6 and G (TcI), TCC1994 (Tcbat), Y and Esmeraldo cl3 (TcII), M6241 cl6 (TcIII), CL Brener (TcVI) Non-Esmeraldo-like (TcIII) and Esmeraldo-like (TcII) haplotypes and homologues from T. c. marinkellei (344), T. dionisii (211), T. rangeli (LDG and AM80) and T. b. brucei (TREU 927). The CATL family signatures of pro-domain motifs ERFININ (ERFN) and GNFD (GTFD) are indicated in bold and underlined, the subsites S1, S2 and S2′ are in bold, and the conserved Trp181 are indicated by (*).The glutamine [Q] of the oxyanion hole, cysteine [C], histidine [H] and asparagine [N] of catalytic triad in the catalytic domain, and 8 cysteines in the C-terminal extension are indicated by arrow heads.