Table 2. The top 10 pairs-gene knockouts or over-expressions that maximize the agronomic properties of the tomato fruit.
| Gene | Gene Annotation | Efficiency 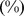 1
1
|
| Acceptability | ||
| LE33G22; LE28J07 | Adenylate kinase, putative; vesicle-associated membrane protein, putative | 16.54 |
| LE15D09; LE33G22 | Vesicle-associated membrane protein, putative; adenylate kinase, putative | 16.54 |
| LE17M21; LE33G22 | Selenoprotein O, putative; adenylate kinase, putative | 16.40 |
| LE17D17; LE33G22 | F-box family protein; adenylate kinase, putative | 16.37 |
| LE15E19; LE33G22 | Ribosomal protein; adenylate kinase, putative | 16.07 |
| LE7I21; LE33G22 | Proline-rich cell wall protein-like; adenylate kinase, putative | 15.90 |
| LE33G22; LE23K21 | Adenylate kinase, putative; amino acid transporter, putative | 15.87 |
| LE33G22; LE2C08 | Adenylate kinase, putative; chloroplast lumen common family protein | 15.85 |
| LE33G22; LE25J09 | Adenylate kinase, putative; AT-HSFA6B, DNA binding/transcription factor | 15.84 |
| LE33G22; LE24D10 | Adenylate kinase, putative; not found | 15.78 |
| Quality (aroma and taste) | ||
| LE27F15; LE29L05 | Protein kinase family protein; branched-chain amino acid aminotransferase | 422.60 |
| LE16D08; LE6G08 | Similar to 60S ribosomal protein L35; sucrose phosphate synthase | 360.63 |
| LE9A08; LE15E23 | GRAM domain-containing protein/ABA-responsive protein-related; putative threonyl-tRNA synthetase | 303.04 |
| LE18E13; LE8A19 | MYB transcription factor; putative glycerophosphoryl diester phosphodiesterase family protein | 263.91 |
| LE32B05; LE4D06 | YABBY2-like transcription factor YAB2; tRNA-dihydrouridine synthase A, putative | 253.33 |
| LE15L08; LE4J06 | Putative rac protein; 50S ribosomal protein L27, chloroplastic | 244.32 |
| LE29E13; LE13F06 | Fyve finger-containing phosphoinositide kinase, fyv1, putative; transmembrane protein, putative | 242.56 |
| LE13F06; LE15J03 | Transmembrane protein, putative; ankyrin-like protein | 240.19 |
| LE17G02; LE15D07 | Pantothenate kinase, putative; polynucleotide kinase- 3′-phosphatase, putative | 239.10 |
| LE15D07; LE20I03 | Polynucleotide kinase- 3′-phosphatase, putative; DEX1, calcium ion binding | 239.03 |
| Quality vs production | ||
| LE13F06; LE15J03 | Transmembrane protein, putative; ankyrin-like protein | 49.79 |
| LE12O13; LE33G22 | Prefoldin subunit, putative; adenylate kinase, putative | 49.16 |
| LE2C24; LE29J02 | ATAB2; RNA binding; GTP-binding protein LepA homolog | 49.15 |
| LE12P11; LE2C24 | Not found; ATAB2; RNA binding | 48.81 |
| LE2C24; LE21J01 | ATAB2; RNA binding; Dolichyl-phosphate beta-glucosyltransferase, putative | 48.28 |
| LE12O13; LE25M06 | Prefoldin subunit, putative; Pre-mRNA-processing protein prp39, putative | 46.63 |
| LE12O13; LE14B20 | Prefoldin subunit, putative; clathrin adaptor complexes medium subunit family protein | 46.18 |
| LE14B20; LE21J01 | Clathrin adaptor complexes medium subunit family protein; dolichyl-phosphate beta-glucosyltransferase, putative | 44.86 |
| LE33B09; LE2C24 | Not found; ATAB2; RNA binding | 44.64 |
| LE18M21; LE14B20 | Cysteine protease; clathrin adaptor complexes medium subunit family protein | 44.05 |
Efficiencies were selected in the RIL where the perturbation maximizes the fruit acceptability, quality and, quality vs production (RILs 103, 142, and 135, respectively).
Knockout genes were showed in bold type and the others were gene over-expressed.
