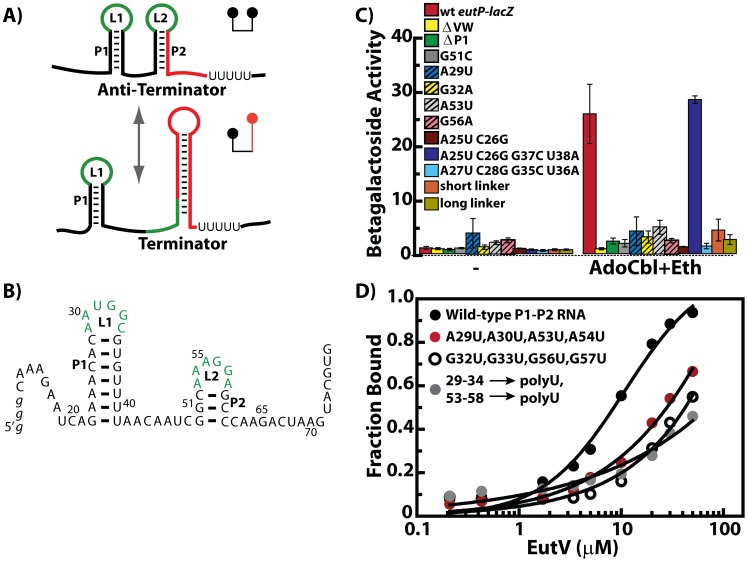
Figure 3. The EutV ANTAR regulator specifically recognizes a dual hairpin RNA motif.
A) Alternate structures formed by the conserved RNA motif are shown. L1 and L2 denote the two terminal loops identified herein and predicted to be involved in ANTAR recognition (green). Altered base pairing allows formation of an alternate RNA structure that includes a terminator stem-loop (red). B) The primary sequence and secondary structure is shown for the dual hairpin RNA motif from the eutP 5′ leader region. Numbering reflects the transcription start site as +1. C) In vivo analysis of RNA mutants using lacZ reporter fusions to the eutP 5′ leader region is summarized as bar graphs. Deletion of eutVW (yellow) abolished induction of lacZ as compared to the wt (red). Deletion of the P1 stem-loop (green) or a mutation in the P2 stem-loop (grey) also negatively affected eutP inducibility. Mutation of the first (adenine) or fourth (guanine) nucleotide within the hexanucleotide loops L1 (blue stripes, yellow stripes) and L2 (grey stripes, pink stripes) significantly decreased eutP induction. Mutations affecting the P1 stem structure (brown) decreased induction but induction could be regained with compensatory mutations that restored the stem structure (blue). Mutations in the sequence of the closing base-pairs of stem-loop P1 abolished induction, even while maintaining the secondary structure (light blue). An increase or decrease in the length of the linker separating P1 and P2 (orange, light green) negatively affected induction. D) Binding isotherms derived from electrophoretic mobility shift assays (EMSA) are shown. Fractional saturation is plotted against protein concentration. EutV (unphosphorylated) bound the eutP 5′ leader region with an apparent K D of 10 µM (black). Binding was significantly deceased in an RNA mutant where the hexanucleotide terminal loops were mutated to uridines (grey). Binding was significantly weaker with RNAs mutated in the first (red) and fourth (open circle) positions of the terminal loops. See also Figure S1 for more information on the seed alignment that was used to derive the predicted RNA secondary structure and Figure S4 for information on EutV purification.