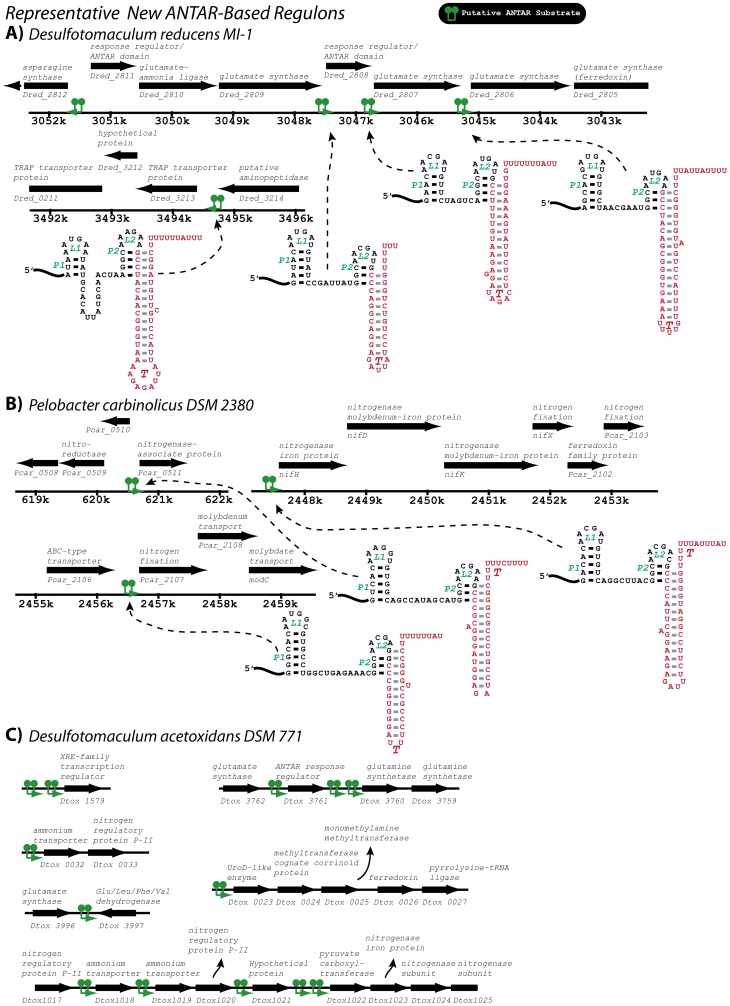
Figure 9. A few representative ANTAR-based regulons identified in this study.
RNA hits (green) from the covariance searches (Tables S1, S2; Figure S2) are shown within their genomic contexts for two representative organisms. Genes are shown along with their annotations (black). The putative ANTAR substrate RNAs appear to be present in multiple operons in the same bacterium, and are thereby likely to participate in control of ANTAR-based regulons. In these examples, the regulons are predicted to be functionally related to control of glutamate metabolism and nitrogenase expression, respectively. See also Figures S1, S2 and Table S1 for more information on the covariance search results. To highlight the sometimes extensive utilization of ANTAR target RNA motifs for certain organisms, the newly identified putative ANTAR-based regulon is shown for Desulfotomaculum acetoxidans. Based on our search this organism utilizes at least 13 ANTAR-based transcription attenuation systems, affecting a total of six transcriptional units that are involved in various aspects of nitrogen metabolism.