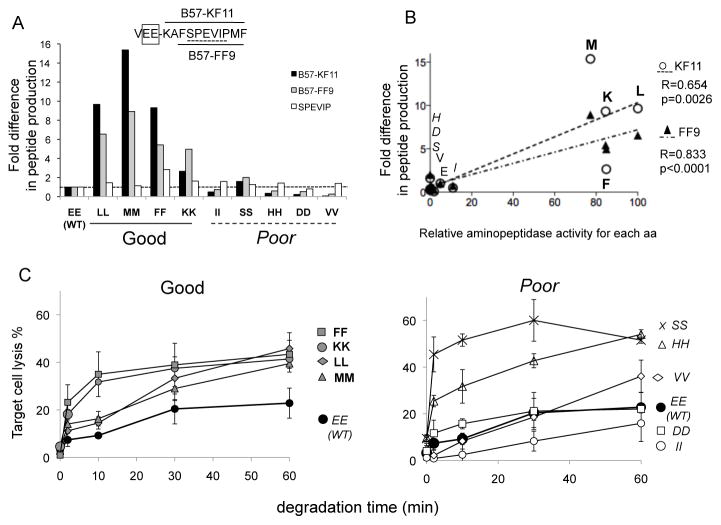
Figure 2. Flanking residues drive the amount and antigenicity of HIV peptides produced in PBMC cytosol.
A. 3-extended KF11 peptides (KAFSPEVIPMF; aa 30–40 in p24) with double mutations flanking KF11 (WT VEE or mutants LL, MM, FF, KK, II, SS, HH, DD, VV) were degraded for 1h in PBMC cytosol. Cleavable motifs are bolded and poor substrates are italicized. Peptides KF11 (black bars), FF9 (grey bars) and SPEVIP (white bars) were identified by mass spectrometry. The intensity of the peak of the 3 peptides was compared between WT and mutants. Representative of 3 experiments. B. Correlation between the fold increase production of KF11 (open circles) and FF9 (black triangles) and the relative PBMC aminopeptidase-specific hydrolytic activity of each amino acid. C. Antigenicity of trimmed KF11 peptides. Degradation products of each extended KF11 peptides preceded with WT or cleavable motifs (left panel: WT EE black circles, FF: grey squares, KK grey circles, LL grey diamonds, MM grey triangles), or poorly cleavable motifs (right panel: WT EE black circles, SS: x, HH: open triangles, VV open diamonds, DD open squares, II: open circles) were purified at different time points, pulsed onto HLA-B57+ B cells used as targets in cytolysis assay with KF11-specific CTL. Average of 3 experiments.