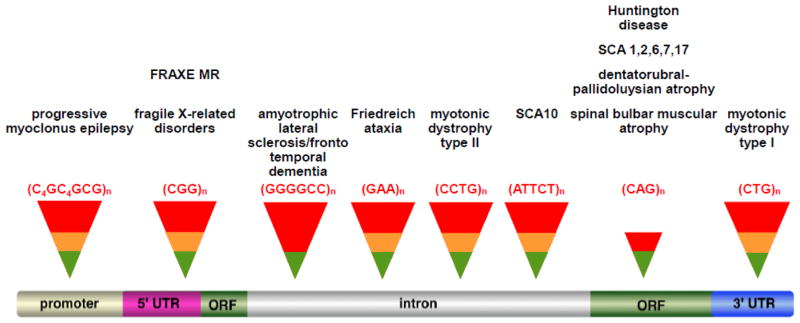
Fig. 1. Diagrammatic representation of the Repeat Expansion Diseases showing the location of the disease-causing repeat on a generic gene.
(see [1] for a more comprehensive description of these diseases). The diseases shown here arise from pathology that is known to be due either to the presence of the repeat in the open reading frame where it generates a polyglutamine tract in the protein that is toxic or in the non-coding portion of the gene where it can affect gene expression in a variety of ways. Three REDs are not shown, Spinocerebellar ataxia type 8 (SCA8), SCA12 and HD-like 2 (HDL2). The pathological effect in SCA8 is thought to be the result of a combination of having the repeats in the coding sequence of one transcript and in the non-coding region of a transcript synthesized in the antisense direction [136]. SCA12 results from a CAG•CTG-repeat in the 5′ region of the PP2R2B gene. However, there is no evidence that expansion results in polyglutamine production and the mechanism responsible for disease pathology is unknown [137]. In the case of HDL2, the disease is caused by a CTG•CAG expansion mutation in a variably spliced exon of junctophilin-3 in the CTG orientation [138]. This seems to exclude polyglutamine tract as the cause of disease pathology in this disorder as well. However, the source of the pathology remains unclear.