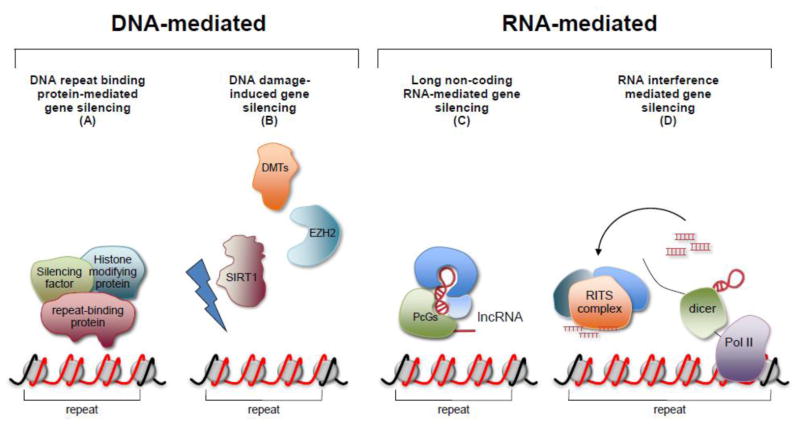
Fig. 3. Four models for repeat-induced gene silencing in FRDA and FXS.
(A) The repeats in the chromosome may act as silencers by binding sequence-specific or structure-specific proteins that then recruit components of the silencing machinery. (B) DNA damage within the repeat may result in the recruitment of the deacetylase SIRT1, EZH2, a component of the repressive Polycomb group (PcG) complexes, and DNA methyltransferases. (C) The repeats in the chromosome may be targeted by PcGs directed to the locus by long non-coding RNA (lncRNA) [141] acting in cis or trans. (D) Silencing may occur via an RNA Interference based mechanism [134] with the long hairpins formed by RNA containing these repeats or duplexes formed by the sense and antisense transcript produced from both of these loci as the source of dsRNA. DMTs: DNA methyltransferases.; lncRNA: long non-coding RNA; PcGs: Polycomb Group Complexes; RITS: RNA-induced transcriptional silencing complex.