FIGURE 3.
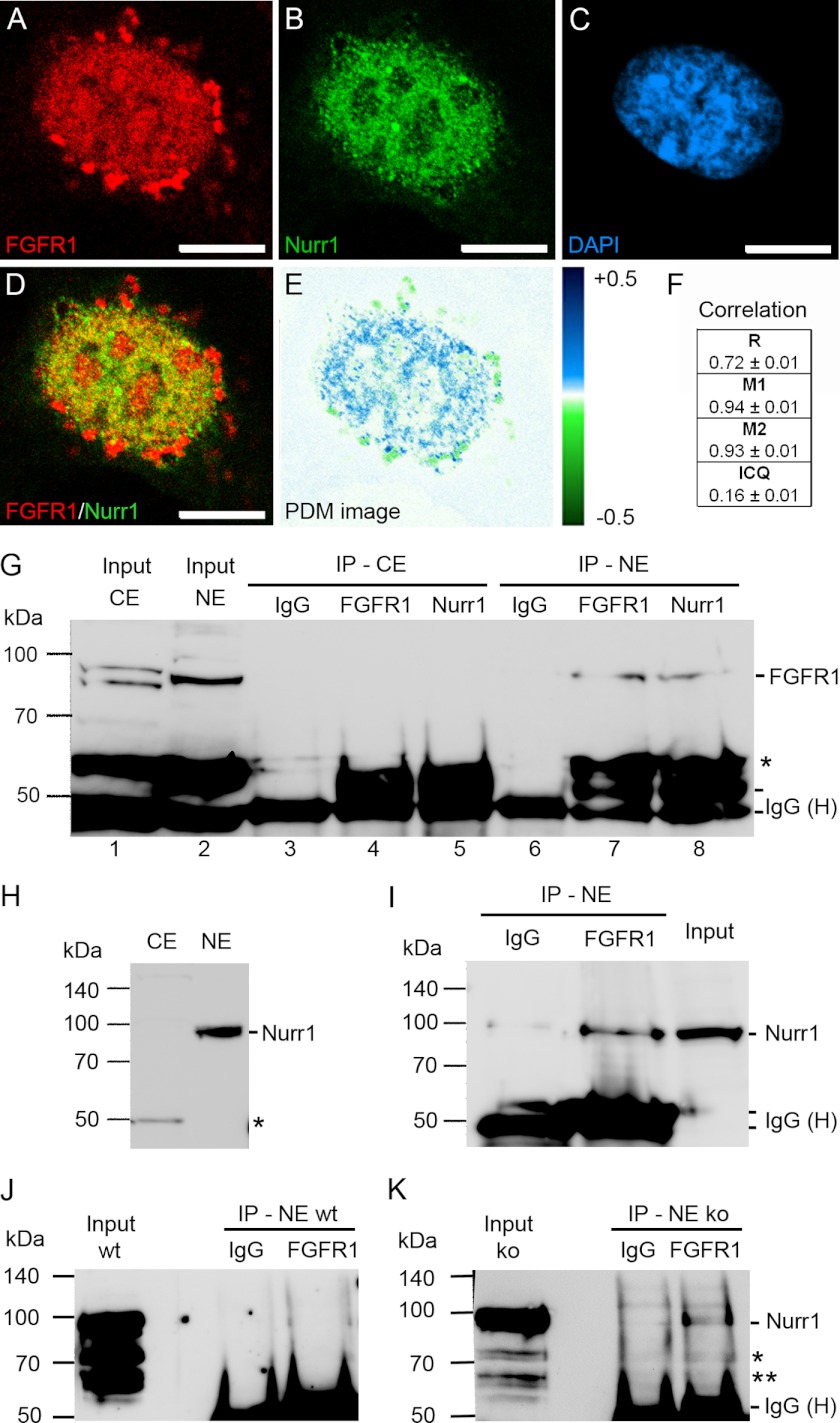
Presence of FGFR1 and Nurr1 in the same nuclear protein complexes of ventral midbrain NPCs. A–F, co-localization analysis of SV40-VM-NPCs cultivated for 24 h in serum-free N2 medium. Confocal images showed granular distribution of FGFR1 (A, red) and Nurr1 (B, green) in the DAPI-stained nucleus (C, blue) of mDA progenitors. FGFR1 and Nurr1 showed co-localization in the nucleus as demonstrated in overlap (D) and PDM images (E, lut; blue, positive PDM and green negative PDM). F, co-localization analysis values with R, Mander's overlap coefficient (red:green pixel ratio = 1.00 ± 0.03); M1, Mander's co-localization coefficient for FGFR1; M2, Mander's co-localization coefficient for Nurr1; and ICQ. Scale bar, 5 μm. G–K, the IP with IgGs represented the negative control for co-precipitation. The input represented the loading control of 100 μg of pure denaturized nuclear protein extract free of denaturized IgG heavy chains (IgG (H)), which are present in IP lanes. Distinct nuclear (NE) ∼90-kDa and cytoplasmic (CE) ∼85- and ∼95-kDa bands of FGFR1 represented different glucosylation forms of the receptor and demonstrated the lack of cross-contamination between fractions (G, lane 1; *, truncated form of FGFR1). The precipitation with anti-Nurr1 resulted in co-precipitation of FGFR1 in the nuclear fraction (G, lane 8). Precipitation of nuclear FGFR1 with anti-FGFR1 is shown as a positive control (G, lane 7). The ∼90-kDa Nurr1 band may result because of post-transcriptional sumoylation of Nurr1 (59) and was present in the nuclear fraction but absent in the cytoplasmic fraction as determined by Western blot assay (H; *, antibody may recognize an additional splice isoform of Nurr1 (17)). Therefore precipitation was performed only in the nuclear fraction, which resulted in co-precipitation of Nurr1 with FGFR1 antibody (I). Precipitation of nuclear FGFR1 in lysates of VM from E14.5 FGF-2 knock out embryos resulted in co-precipitation of Nurr1 (K), whereas in IPs of VM lysates from E14.5 wild-type embryos, the signal was not clearly detectible (J; * and **, unmodified and/or splice forms of Nurr1). The amount of used material was limited by the availability of fresh tissue.