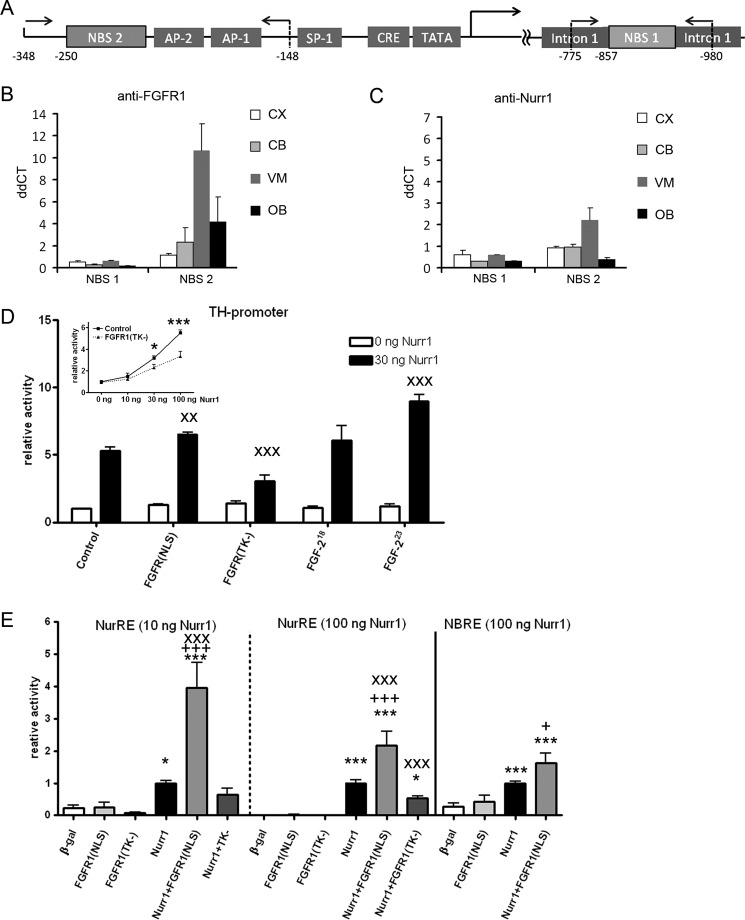
FIGURE 6.
Nurr1 and FGFR1 bind to and cooperatively activate transcription from TH promoter. A, schematic of the TH gene promoter region. Binding sites for other factors are labeled accordingly. Arrows with numbers indicate positions of primers for ChIP. B and C, ChIP was performed with a panel of antibodies against FGFR1, Nurr1, and control IgG with subsequent quantitative PCR analyses of selected potential NBS on TH. IgG was used as a negative control. Graphs show ΔΔCt means ± S.E. of triplicate samples. CX, cortex; CB, cerebellum; VM, ventral midbrain (containing substantia nigra region); OB, olfactory bulb. D, co-transfection of human neuroblastoma cells with Nurr1–3×FLAG showed a dose-dependent increase of TH promoter-dependent luciferase reporter gene expression. The Nurr1-mediated TH promoter activity was significantly diminished by co-transfection of the dominant-negative form of FGFR1 lacking the tyrosine kinase activity [FGFR1(TK−)] starting at 30 ng of Nurr1-FLAG (inset). Co-transfection of FGFR1(NLS), FGFR1(TK−), as well as FGF-223 with Nurr1-FLAG, resulted in significant interaction altering Nurr1-dependent TH promoter activation. The noninteracting isoform FGF-218 did not influence TH promoter activity mediated by Nurr1. Two-way ANOVA on interaction with Nurr1: x, p < 0.05; xx, p < 0.01; xxx, p < 0.001. E, co-transfection of FGFR1(NLS) in neuroblastoma cells enhances Nurr1-NurRE and -NBRE-dependent luciferase transcription. The dose-dependent effects of NurRE activation by 10 and 100 ng of co-transfected Nurr1-FLAG are significantly potentiated by FGFR1(NLS), whereas the co-transfection of FGFR1(TK−) significantly inhibits 100 ng of Nurr1-NuRE activation. Co-transfection of FGFR1(NLS) also enhances Nurr1-dependent transcription from NBRE motive. One-way ANOVA significance to β-galactosidase (β-gal) is expressed as * and to Nurr1 as +. Two-way ANOVA displays Nurr1 interactions as x. Significance levels: *, p < 0.05; **, p < 0.01; ***, p < 0.001.