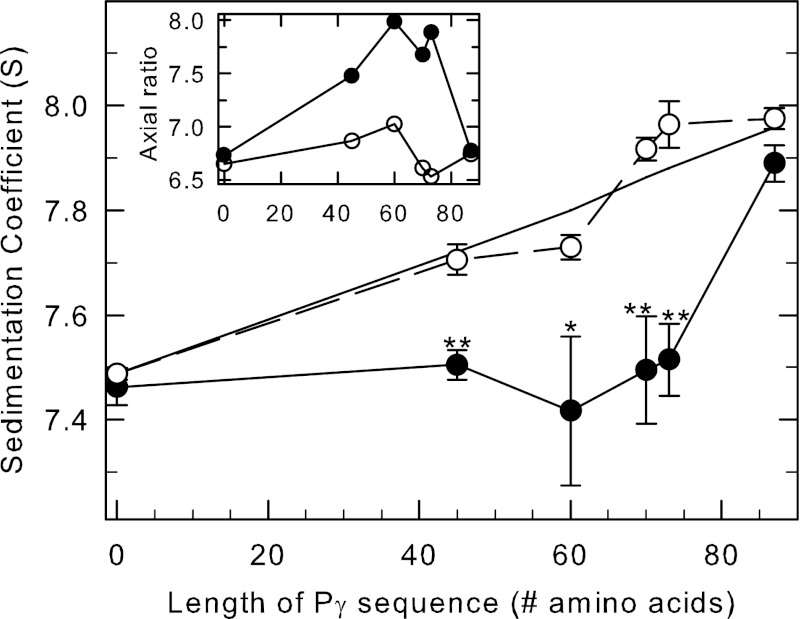
FIGURE 4.
cGMP-induced conformational changes in PDE6 observed when Pγ truncation mutants are bound to the catalytic dimer. Purified, nucleotide-depleted Pαβ-IAF (10 nm) was incubated with 1 μm concentrations of the following: nothing (control), full-length Pγ (Pγ(1–87)), and the C-terminal truncation mutants Pγ(1–45), Pγ(1–60), Pγ(1–70), and Pγ(1–73). One portion of each preparation was incubated with 10 mm cGMP, and samples were centrifuged as soon as possible thereafter to minimize cGMP hydrolysis. The x axis represents the number of amino acids in each of the C-terminal truncation mutants. Data points represent the mean ± S.E. of 4–9 separate experiments. The asterisks indicate statistical difference at p < 0.05 (*) p < 0.005 (**) level of significance. The straight line is the predicted s value for each sample, calculated using the additional mass contributed by each Pγ truncation mutant, assuming a binding stoichiometry of 2 Pγ per Pαβ and no change in shape of the reconstituted protein. Inset, the axial ratio was calculated with SEDNTERP based on the composition molecular weight and the s value for each condition in the absence (open circles) and presence (filled circles) of cGMP.