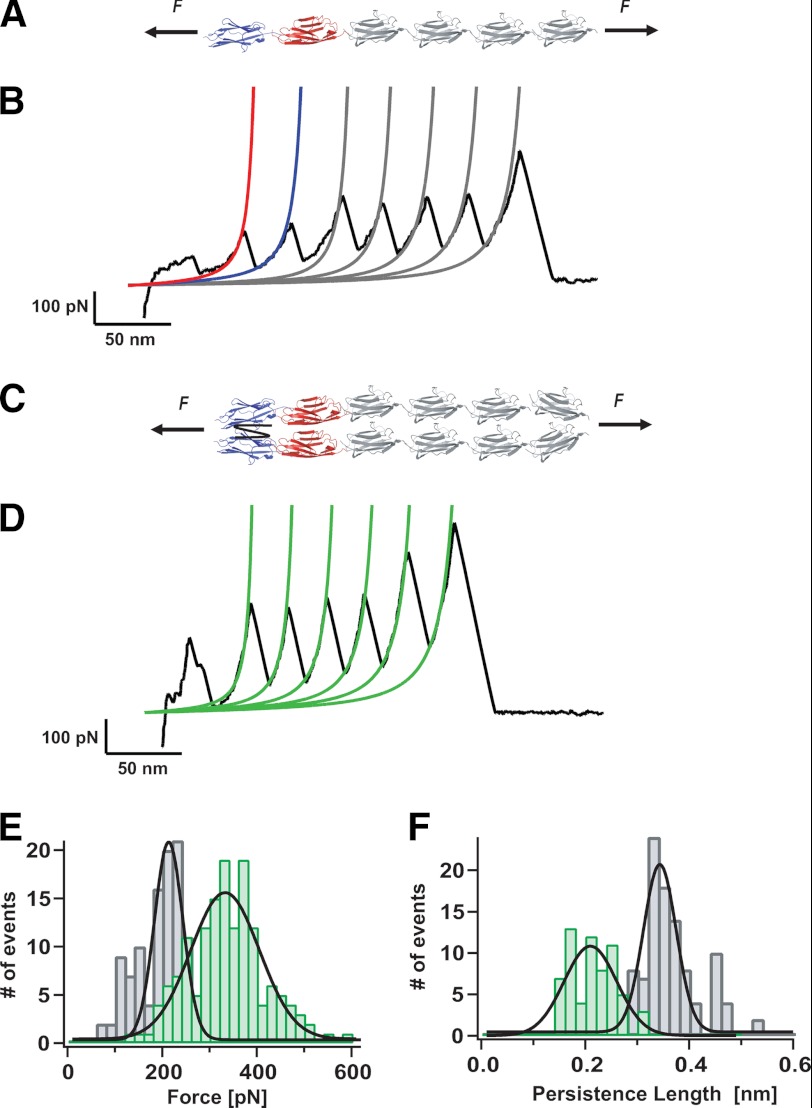
FIGURE 3.
Mechanical fingerprint of a parallel dimeric 2((I27)4Z1Z2) polyprotein. A, scheme of the engineered (I27)4Z1Z2 polyprotein under mechanical unfolding conditions. B, typical unfolding trajectory obtained form pulling a (I27)4Z1Z2 monomer. Fitting the WLC model of polymer elasticity to the traces (red fit for Z1, blue fit for Z2, and gray fits for I27 modules) measures the increment in contour length, ΔL, and the persistence length, P, for the monomeric species. C, diagram of the protein dimer 2((I27)4Z1Z2). D, typical unfolding trace corresponding to in-register unfolding of the 2((I27)4Z1Z2) dimer. Fittings to the WLC (green fits) measure the ΔL and P values associated to each unfolding peak. E, histogram corresponding to the unfolding forces measured for the protein monomer trajectories (gray bars) and for the protein dimers (green bars). The measured average unfolding for the dimers (338 ± 85 pN) is almost double that obtained for the monomers, 189 ± 48 pN, n = 107. F, by contrast, the persistence length is almost halved for the protein dimers (0.21 ± 0.04 nm, n = 63) with respect to that obtained for the protein monomers (P = 0.36 ± 0.06 nm).