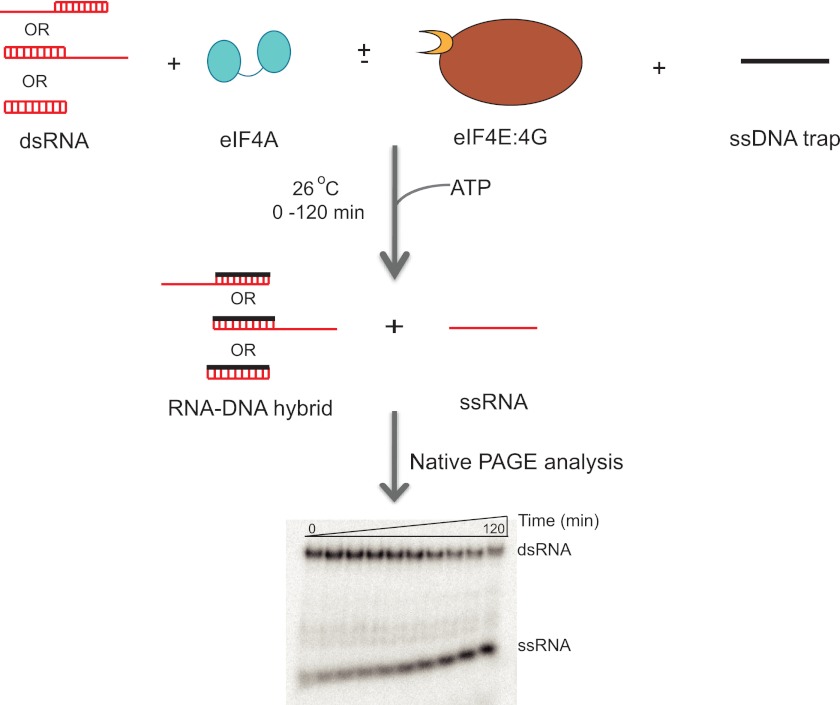
FIGURE 3.
Schematic representation of the RNA unwinding assay. A flow chart of the RNA unwinding assay. Briefly, the dsRNA substrate (10 nm) was incubated with eIF4A (100 nm, with or without 300 nm eIF4E·eIF4G) and DNA trap (3 μm). The DNA trap has the same sequence as the labeled RNA oligonucleotide (top strand of duplexes in red) and serves to prevent reannealing of the two RNA strands after unwinding. The unwinding reaction was initiated by the addition of ATP (3 mm). The time course (0–120 min) was monitored at 26 °C and analyzed by native PAGE cooled with a circulating water bath. A representative unwinding gel is shown here. The gel was scanned and quantified using a PhosphorImager. As with the RNA binding and ATPase, the reaction was carried out in Recon buffer.