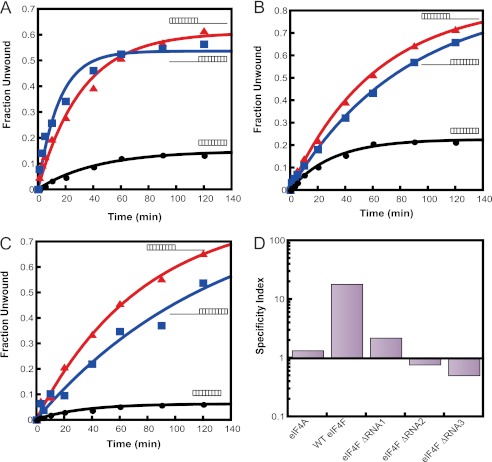
FIGURE 7.
Substrate specificity of mutant eIF4F complexes. The RNA unwinding activities of the eIF4F ΔRNA mutants were measured with three different substrates (Table 1). Representative time courses of unwinding are shown with the different RNA duplexes catalyzed by eIF4F ΔRNA1 (A), eIF4F ΔRNA2 (B), and eIF4F ΔRNA3 (C). The substrates are schematically represented next to each time course. The unwinding rate constants (Table 3) were obtained by fitting each plot to a first-order exponential equation. D, specificity index, defined as the ratio of the unwinding rate constant for the duplex with the 30-nt 5′-overhang to that of the duplex with the 30-nt 3′-overhang, for each eIF4F complex. A specificity index >1 indicates a preference for unwinding the duplex with the 5′-overhang and a value <1 indicates a preference for the duplex with the 3′-overhang. Conditions were as in Fig. 3, except in experiments with eIF4F ΔRNA1 500 nm eIF4E·eIF4G was used.