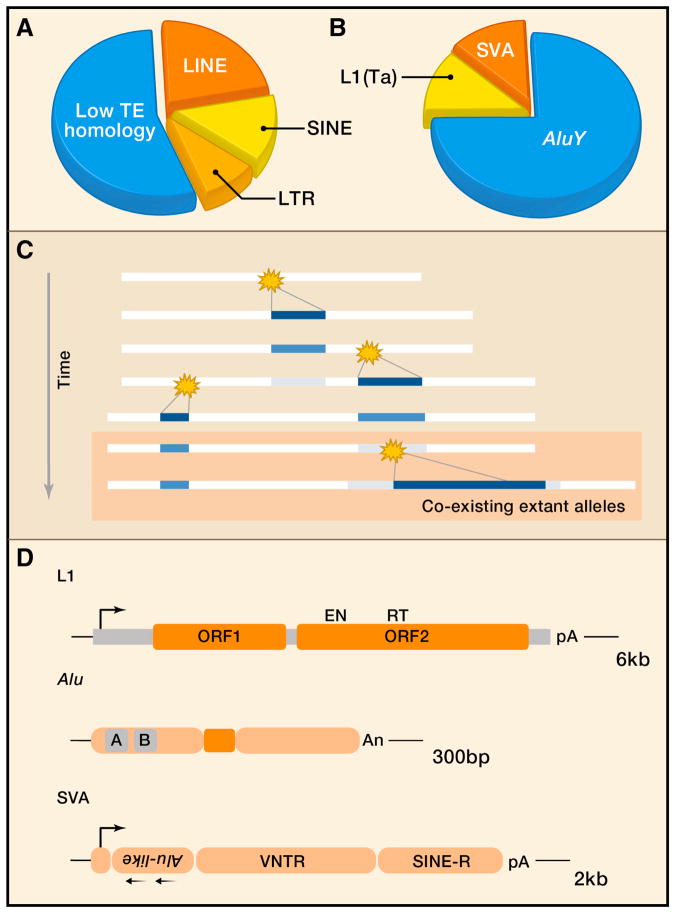
Figure 1.
Human retrotransposons. A. Composition of the human genome with respect to high copy number repeats. Data are from the RepeatMasker analysis of the hg19 human genome assembly (Genome Reference Consortium GRCh37). The illustration shows fractions of the genome derived from the major orders (Wicker et al., 2007) of retrotransposons. The remaining 55% of the genome bears low homology to TEs, although substantial portions may be derived from mobile DNA. B. Transposon types illustrated as fractions of total ongoing activity. AluY are the most prolific source of new insertions at one de novo germ line insertion per 20 births; L1 and SVA are thought to be comparable in current activity, responsible for one insertion per 100–200 births. C. Schematic showing an accumulation of interspersed repeat insertions over time. New integrations are stochastic events in individuals (star), such that co-existence with the antedating ‘empty’ allele occurs in the population initially. In the schematic, two alleles are present currently, reflecting presence and absence of the most recent insertion [a retrotransposon insertion polymorphism (RIP)]. As persisting insertions age, they become ‘fixed’ or invariant in the population and decrease in sequence likeness to similar elements (black to gray). D. Structure of the most active retroelements in human genomes. L1 LINEs have a CpG rich 5′UTR with an internal RNA polII promoter (5′ rightward arrow), two open reading frames encoding ORF1p and ORF2p (pink segments), and a 3′ UTR with a polyadenylation (pA) sequence. The ORF2 reading frame encodes endonuclease (EN) and reverse transcriptase (RT) domains. Alu elements are derived from 7SL ribosomal RNAs; they have two internal ‘monomer’ sequences with a centrally located A-rich sequence and an RNApolIII promoter (A and B, gray). The sequence ends in multiple adenosines (An). SVAs are composites of other repeats, from left to right, a CCCTCTn repeat, two tandem Alu-like sequences in antisense (leftward arrows), a VNTR (variable number tandem repeat) region, and a SINE-R region with HERV homology. Sequence suggests RNApolII driven transcription (5′ rightward arrow), and there is a 3′ AAUAAA sequence (pA).