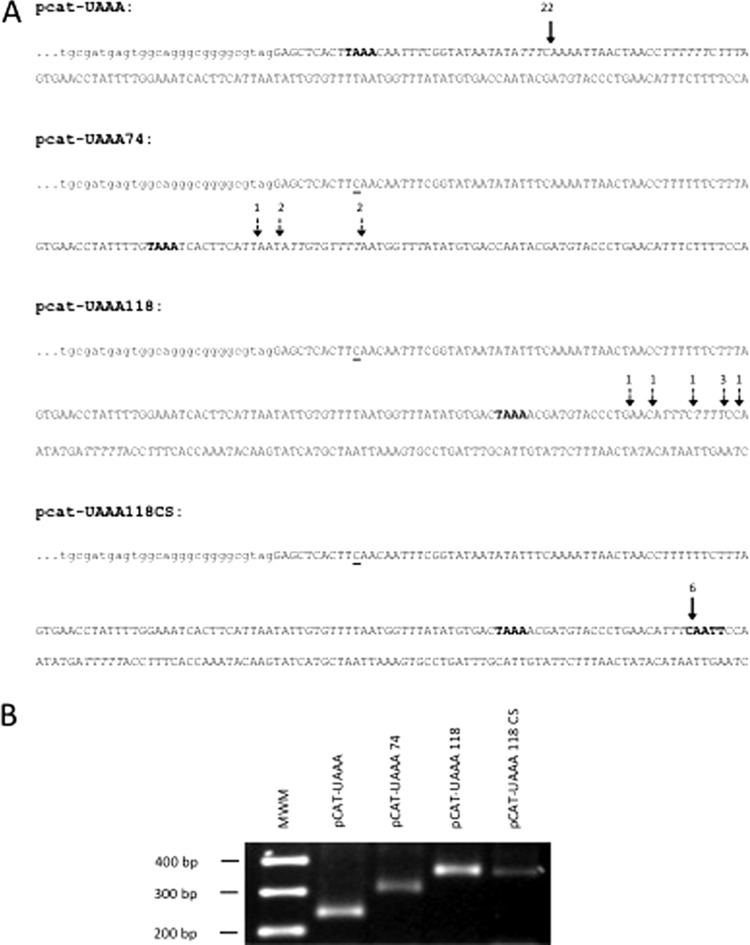
Fig 5.
Relocation of the UAAA motif redirects the polyadenylation site. (A) DNA sequences of the plasmid constructs derived from the pcat-UCAA mutant, in which the UAAA motif was relocated downstream from the parental site. pcat-UAAA, parental sequence. The arrow denotes the site where all mRNAs were found to be polyadenylated. pcat-UAAA74, the UAAA motif (bold) was placed 74 nt downstream from the stop codon, and the parental UAAA motif was mutated (underlined C). The arrows represent the close-by sites where mRNAs were found to be polyadenylated. pcat-UAAA118, the UAAA motif (bold) was placed 118 nt downstream from the stop codon, and the parental UAAA motif was mutated (underlined C). The arrows represent close-by sites where mRNAs were found to be polyadenylated. pcat-UAAA118CS, the CAATT motif (bold), proposed as one of the cleavage site consensus motifs, was placed 20 nt downstream from the UAAA sequence in the pcat-UAAA118 plasmid. The arrow represents the site where all mRNAs were polyadenylated. The numbers above the arrows represent the number of clones found polyadenylated at each position. All polyadenylation sites were inferred from cDNA cloning and sequencing. (B) Evaluation of the size and homogeneity of the cat mRNAs by RT-PCR. Bands of the expected size were obtained, as predicted from the position where the UAAA motifs were introduced. Amplification products were analyzed on a 2.2% agarose gel and stained with ethidium bromide.