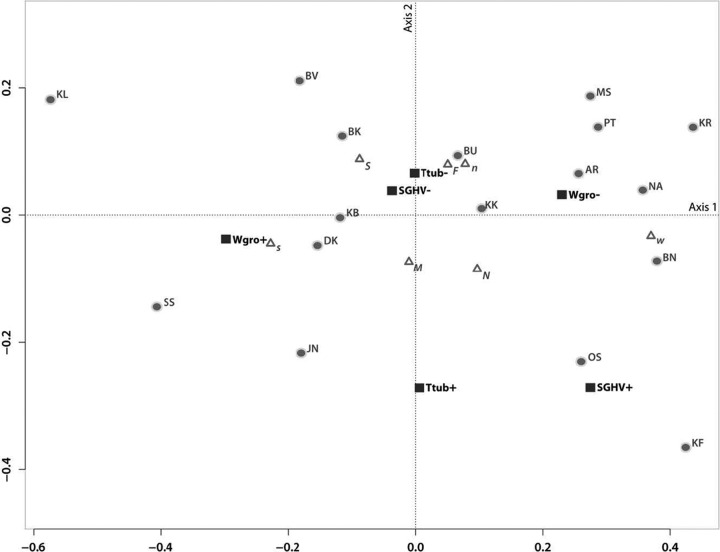
Fig 5.
Multiple correspondence analysis plot. The first two axes, representing 63% of the total variation, are plotted. Eigenvalues (λ1 = 0.038 and λ2 = 0.013) and proportions of explained inertia (τ1 = 46.9% and τ2 = 16.1%) have been corrected using the Greenacre (21) method. Populations (abbreviations from Table 1), microsatellite genotype clusters (n, north; s, south; w, west), mtDNA haplogroups (N, north; S, south), and sex (F, female; M, male) are depicted to show their association with presence (+) or absence (−) of Wolbachia (Wgro), Glossina pallidipes salivary gland hypertrophy virus (SGHV), and Trypanosoma (Ttub). Triangles represent supplementary elements (microsatellite, mtDNA, and sex, plotted post hoc), while infections and populations are denoted with squares and circles, respectively.