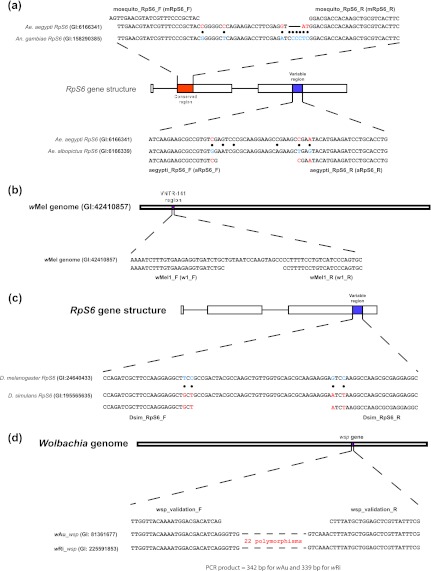
Fig 1.
Development of gene markers to detect and quantify Wolbachia infection in Ae. aegypti and D. simulans. (a) Coding sequences of the RpS6 gene from Ae. aegypti and Anopheles (An.) gambiae were aligned; a conserved region was selected to place the universal Aedes primers mRpS6_F and mRpS6_R. Nucleotide alignments of the coding sequences from Ae. aegypti and Ae. albopictus RpS6 were used to identify a variable region in which to position a pair of Ae. aegypti-specific primers (aRpS6_F and aRpS6_R), with 1 to 2 diagnostic nucleotides at the 3′ end of each primer. (b) The complete genome sequence of Wolbachia wMel was used to design a Wolbachia-specific marker at the VNTR-141 locus (see reference 9). The GenBank identifiers (GI) of the source sequences are given. Solid circles indicate polymorphic sites. (c) The variable region of the coding sequences in the RpS6 gene between Drosophila melanogaster and D. simulans is shown. Primers were designed to amplify D. simulans but not D. melanogaster. Solid circles indicate polymorphic sites. (d) Universal primers were placed at the conserved regions between the wRi and the wAu strains. These primers flank an ∼290-bp highly variable region that contains 22 polymorphic sites. The GI numbers of the source sequences are given. Note: reverse primers (mRpS6_R, aRpS6_R w1_R, Dsim_RpS6_R, and w1_R) are illustrated in the sense direction; their 5′-to-3′ sequences are described in the text.