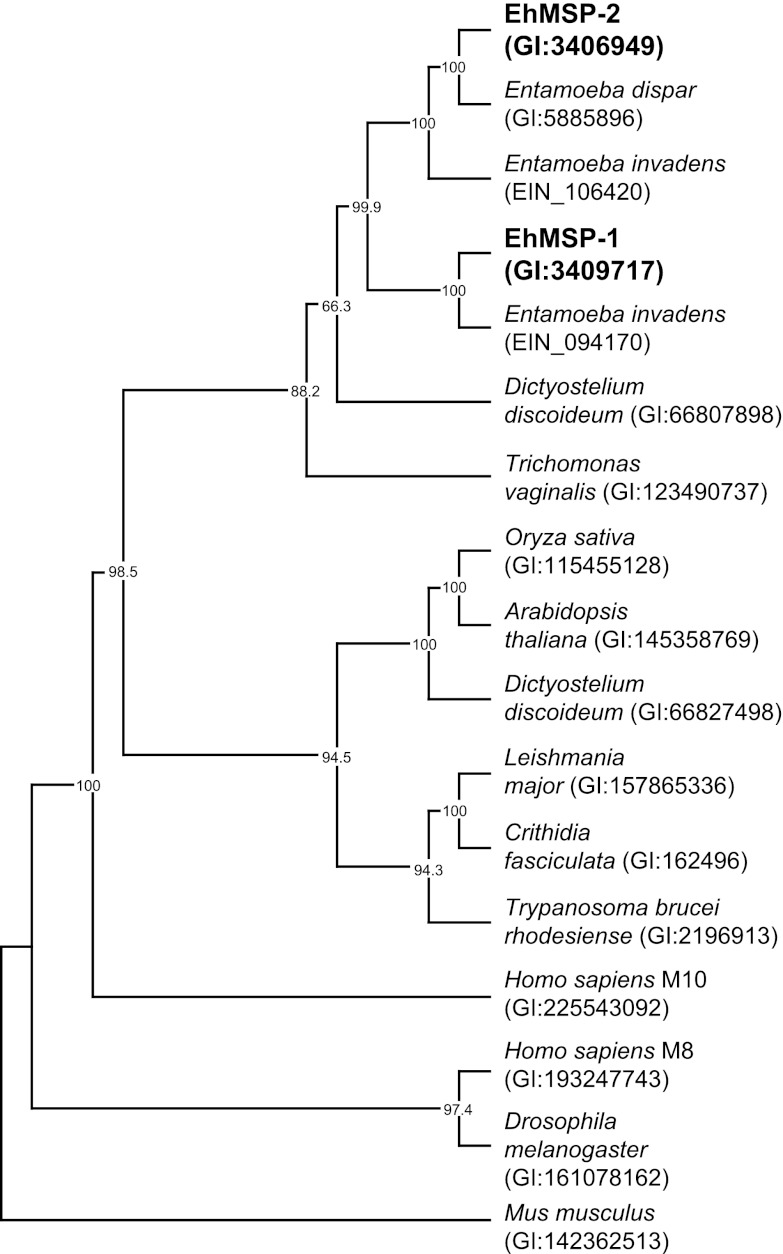
Fig 1.
Phylogenetic analysis of EhMSP-1 and EhMSP-2 suggests that E. dispar has lost the gene encoding EhMSP-1. The predicted amino acid sequences for EhMSP-1 and EhMSP-2 were used to identify genes encoding homologous proteins by pBLAST in E. dispar and E. invadens. The predicted amino acid sequences of these proteins were aligned with those of representative M8 metalloendopeptidases from diverse species and analyzed using maximum parsimony with the program PHYLIP. A human M10 family matrix metalloendopeptidase was included as an outgroup. The consensus tree from 1,000 bootstrap iterations and bootstrap proportions >60% are shown.