Fig 3.
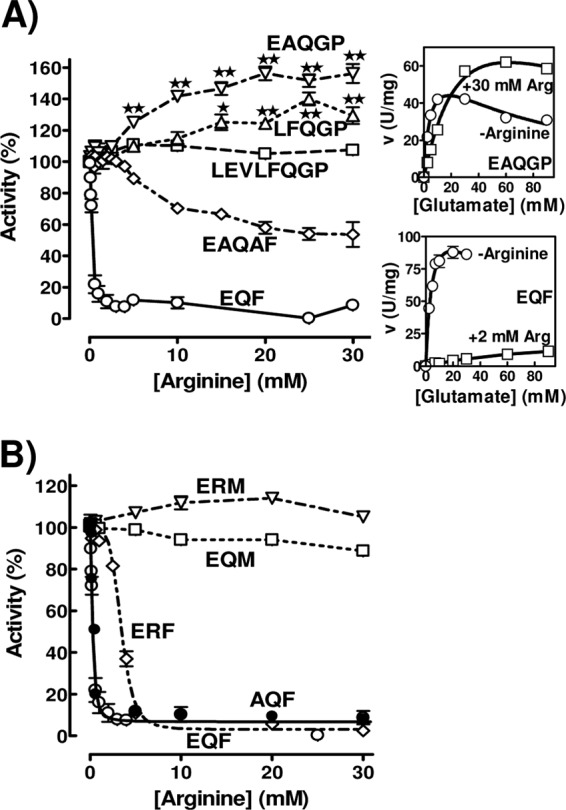
Influence of linker mutations on the effect of arginine on PaNAGS activity (expressed as a percentage of that for each form in the absence of arginine). See Fig. 2C for the key to each mutant form. Mutant forms having (A) or not having (B) a lengthened linker are compared with the wild-type form (EQF). The activation of the EAQGP and LFQGP forms by arginine is statistically significant for the points marked with double stars (P < 0.001) or single stars (P < 0.01) (tested by analysis of variance [ANOVA], followed by the Bonferroni test for individual points; n = 4 to 8). The insets to the right of panel A show the glutamate concentration dependency of the velocity for the form with the EAQGP linker sequence (top inset) and for the wild-type enzyme (bottom inset). The curves drawn in these insets are those for hyperbolic kinetics with substrate inhibition for KmGlu, KIGlu, and Vmax, as follows: 4.9 mM, 69 mM, and 67 U/mg, respectively, for EAQGP without arginine; 37 mM, 103 mM, and 136 U/mg, respectively, for EAQGP with 30 mM arginine; 5.2 mM, 72 mM, and 136 U/mg, respectively, for EQF (wild type) without arginine; and 85 mM, ∞ (no substrate inhibition), and 22 U/mg, respectively, for EQF with 2 mM arginine.
