Fig 6.
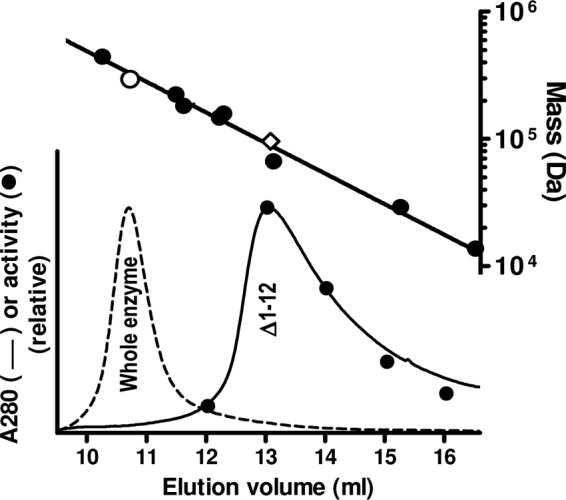
Size-exclusion chromatography of the Δ1–12 form of PaNAGS. The chromatographic profile (continuous line) is compared with that of the wild-type enzyme (broken line), both injected in 0.2-mg amounts. The procedure and system are described in the section on cleavage with PreScission protease and the separation of the two domains in Materials and Methods. In the case of Δ1–12, enzyme activity was measured in 1-ml collected fractions (closed circles) (bottom plot). The upper line is the semilogarithmic plot of the masses of marker proteins (closed circles) versus their elution volumes. The open symbols correspond to the protein peaks below them for the following sequence-deduced masses: 294.4 kDa for the whole enzyme, assuming that it is hexameric (○), and 95.5 kDa for Δ1–12 enzyme form, assuming that it is dimeric (♢). The protein standards used are ferritin (440 kDa), β-amylase (224 kDa), T. maritima acetylglutamate kinase (182 kDa) (28), aldolase (158 kDa), alcohol dehydrogenase (147 kDa), bovine serum albumin (66.4 kDa), carbonic anhydrase (29 kDa), and RNase (13.7 kDa).
