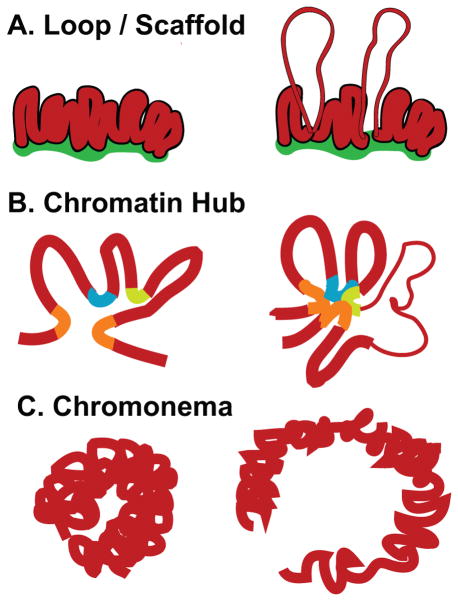
Figure 1. Higher-order chromatin folding models (left) and changes associated with transcriptional activation (right).
(A) Chromatin domain/loop model postulates loops of chromatin anchored by a nuclear matrix with decondensation from 30 nm chromatin fiber to 10 nm nucleofilament accompanying activation. (B) Chromatin hub postulates a 3D conformation is created by interactions between distant cis regulatory elements; changes in transcriptional activation may depend on which cis elements contribute to this hub. (C) Chromonema model postulates folding of chromatin into large-scale chromatin fibers. Transcription is accompanied by a decondensation, elongation, and straightening of these fibers but still occurs within the context of highly condensed chromatin.