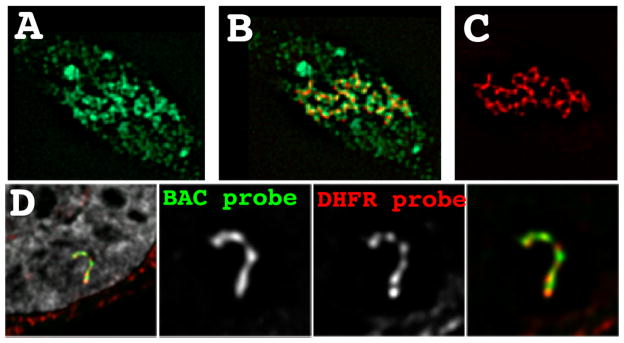
Figure 5. Transcription on a condensed template.
(A–C) Tethering of the VP17 acidic activator using a EGFP-lac repressor-VP16 fusion protein (red) to a heterochromatic amplified chromosome region results in extensive decondensation and extended large-scale chromatin fibers. BrUTP pulse labeling (green) shows large amounts of nascent transcripts surrounding these large-scale chromatin fibers. (D) DHFR multi-copy BAC transgenes assemble into a compact large-scale chromatin fiber despite transcriptional activity within 2–3 fold of the endogenous DHFR gene. Fluorescence in situ hybridization using the entire BAC as a probe (green) versus probes covering the 34 kb DHFR gene (red) reveal the absence of any detectable looping of the DHFR genes outside of these large-scale chromatin fibers. DAPI staining is grey.