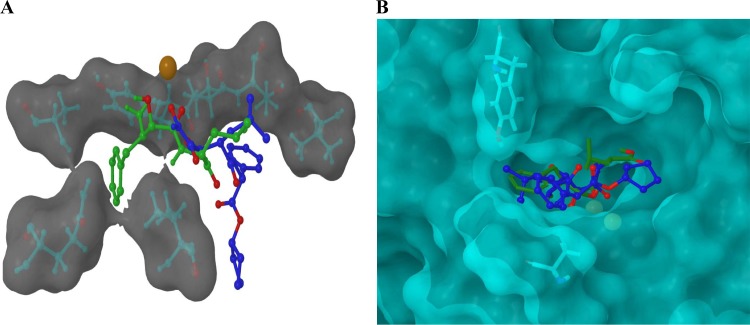
Fig 2.
(A) The calculated locations of CHR-2863 and bestatin within the binding site of rPfM1AAP obtained by molecular docking studies using Glide. CHR-2863 (blue) is predicted to occupy a region of the substrate-binding cavity different from that observed in both the X-ray-derived solid-state structure and our docking analysis for bestatin (green). Oxygen atoms are highlighted in red, the zinc ion is highlighted in orange/brown and the proximal hydrophobic residues are shown inset into a semitransparent Van der Waals (VDW) molecular surface. (B) The calculated locations of CHR-2863 and bestatin within the binding site of rPfM17LAP obtained by molecular docking studies using Glide. Analysis revealed that the phenyl group of CHR-2863 and the isopropyl group of bestatin occupy the same hydrophobic region of the rPfM17LAP protein, while the phenyl group of CHR- 2863 projects into a region proximal to Tyr493 and Ala388. Oxygen atoms are highlighted in red, the zinc ions are highlighted in yellow, and the proximal residues are shown inset into a semitransparent VDW molecular surface.