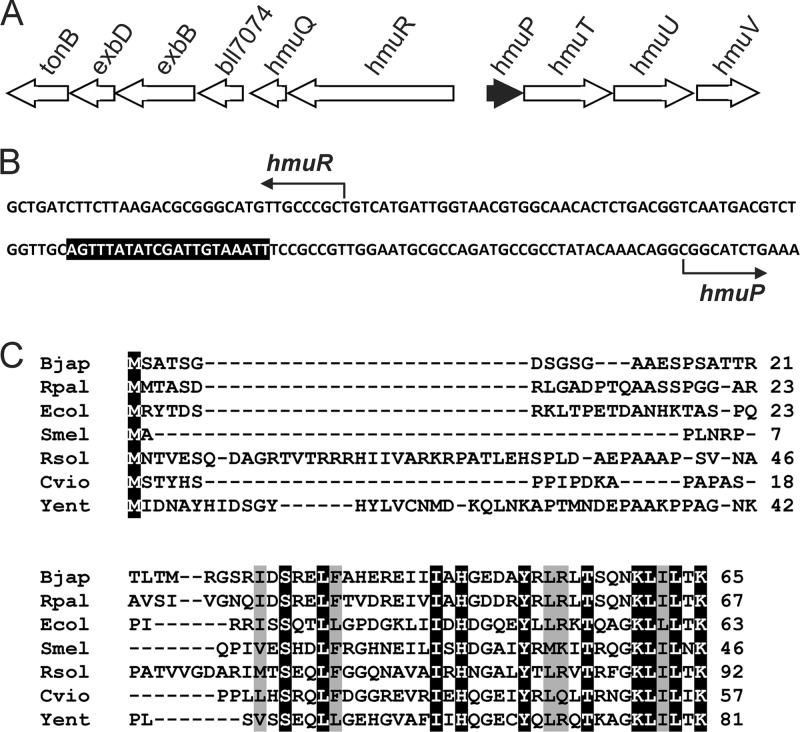
Fig 1.
Localization of hmuP within the heme utilization heme locus and comparison of B. japonicum HmuP with homologs in other bacteria. (A) The hmuP gene was found upstream of the divergent genes hmuR and hmuT in the same orientation as hmuT. (B) Partial nucleotide sequence of the hmuR-hmuP intergenic region. The sequence shown corresponds to the coding strand of hmuP and the template strand of hmuR. The bent arrows show the transcription start sites of hmuR and hmuP as determined by Nienaber et al. (18). The shaded region denotes the ICE. (C) Alignment of HmuP homologs in B. japonicum (Bjap), were compared with those of Rhodopseudomonas palustris (Rpal; YP_782068.1), E. coli (Ecol; YdiE), Sinorhizobium meliloti (Smel; NP_386533.1), Ralstonia solanacearum (Rsol; NP_520087.1), Chromobacterium violaceum (Cvio; NP_903565.1), and Yersinia entercolitica (Yent; HEMP_YEREN). Identical amino acids are shaded in black with white letters, and conserved residues are shaded in gray.