Table 1. Data-collection and refinement statistics for ADAM-8–batimastat (PDB entry 4dd8).
Values in parentheses are for the last shell.
| Data-collection statistics | |
| Reservoir solution | 0.1 M Tris pH 8.0, 1 M sodium formate, 10% methanol |
| Space group | P21 |
| Unit-cell parameters | |
| a (Å) | 91.6 |
| b (Å) | 50.9 |
| c (Å) | 93.5 |
| α (°) | 90 |
| β (°) | 102.4 |
| γ (°) | 90 |
| Resolution range (Å) | 50–2.1 |
| No. of observations | 176092 |
| No. of unique reflections | 49479 |
| Average multiplicity | 3.6 (2.8) |
| Completeness (%) | 99.3 (95.9) |
| Rmerge† (%) | 10.6 (36.5) |
| Average I/σ(I) | 10.9 (2.9) |
| Refinement statistics | |
| Resolution (Å) | 44.7–2.1 (2.18–2.10) |
| No. of reflections | 47689 |
| Rcryst‡ (%) | 18.8 |
| Rfree§ (%) | 25.5 |
| No. of non-H atoms | |
| Protein | 4016 |
| Ligand | 128 |
| Ions | 22 |
| Solvent | 415 |
| Mean B factor (Å2) | |
| Protein (molecule A, B, C, D) | 23.32, 22.43, 24.21, 26.25 |
| Ligand (molecule A, B, C, D) | 25.40, 26.41, 23.43, 27.72 |
| Ions | 27.55 |
| Solvent | 26.32 |
| R.m.s. deviations from ideal values | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 0.682 |
| Ramachandran statistics¶ | |
| Most favored regions (%) | 97.5 |
| Disallowed regions (%) | 0.2 |
R
merge = 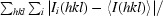
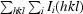 , where I
i(hkl) is the intensity of the ith observation and 〈I(hkl)〉 is the mean intensity of the reflection.
, where I
i(hkl) is the intensity of the ith observation and 〈I(hkl)〉 is the mean intensity of the reflection.
R
cryst = 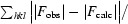
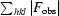 calculated from the working data set.
calculated from the working data set.
R free was calculated from 5% of data randomly chosen not to be included in refinement.
The Ramachandran results were determined by MolProbity.
