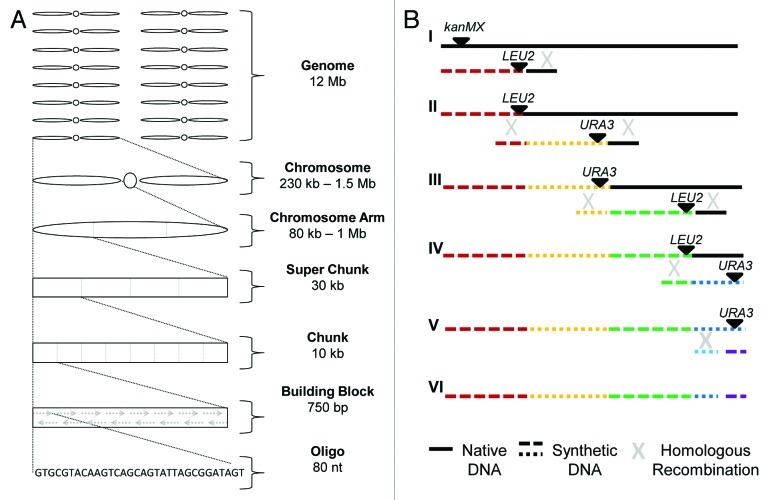
Figure 1. Genome modularity and integration of synthetic DNA. (A) The yeast genome is subdivided into increasingly smaller segments to facilitate construction and assembly of the synthetic Sc2.0 genome (not to scale). The assembly pipeline may be entered from multiple points. The assembly technique utilized by Build-a-Genome students begins at the bottom of the assembly pipeline, constructing building blocks from oligos.9 Commercially synthesized DNAs used to construct semi-synVIL were obtained as chunks, and assembled into a super chunk prior to integration in the yeast genome. SynIXR entered the pipeline as a chromosome arm. (B) Synthetic DNA is iteratively integrated into the yeast genome to replace native DNA (not to scale). The native chromosome, marked with kanMX, is targeted for replacement by integration of synthetic DNA, marked with LEU2. An “endcap” directs homologous recombination to the region/s flanking the synthetic sequence. The resulting Leu+ G418S semi-synthetic chromosome is then targeted for replacement by integration of a URA3-marked synthetic DNA fragment (II). Iterative transformations with synthetic DNA fragments alternately marked with LEU2 and URA3 sequentially replace the native DNA (III-IV). The final URA3 marker is replaced by transformation with synthetic sequence lacking the URA3 gene and subsequent selection on 5-FOA (V) to generate the complete unmarked synthetic chromosome.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.