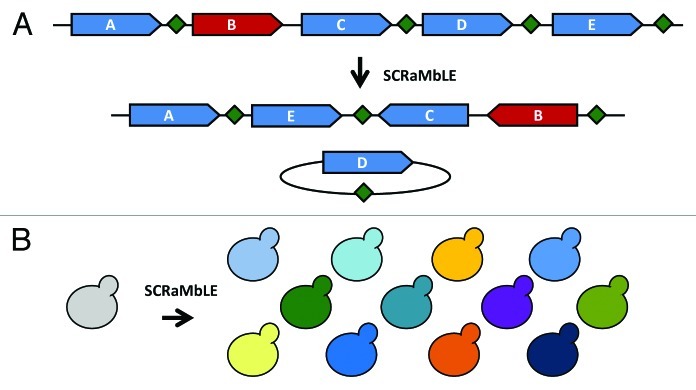
Figure 2. SCRaMbLE restructures the synthetic genome. (A) LoxPsym sites (green diamonds) are inserted in the 3′UTR of each non-essential gene (blue arrows); essential genes (red arrow) do not have an associated loxPsym site. The symmetry of loxPsym sites permits both translocations/inversions and deletions at each site. Complex rearrangements result from induction of SCRaMbLE. In the example shown, genes “B” and “C” are inverted, “E” has been excised and reintegrated, and “D” has been lost from the SCRaMbLEd chromosome. (B) Induction of SCRaMbLE in a synthetic strain (gray) results in a significant increase in genetic diversity (colors). Following selection for a desired phenotype, which can range from simple viability to increased ability to produce a desirable substance, genome content and structure of SCRaMbLEd strains can be analyzed by PCRTag analysis,7 comparative genome hybridization (CGH), molecular karyotyping and/or whole-genome sequencing.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.