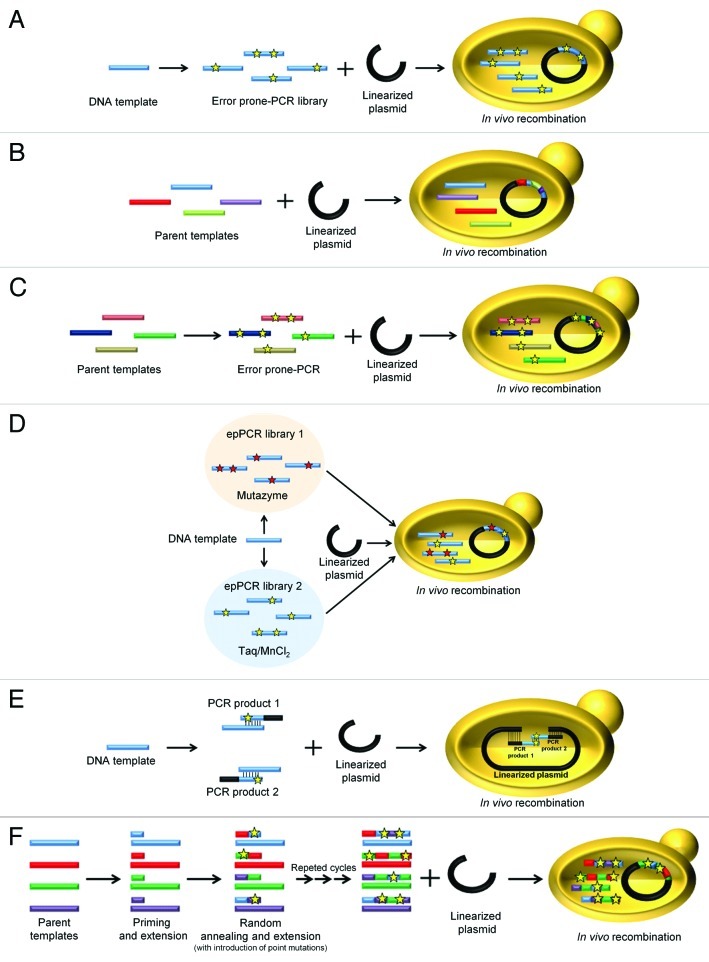
Figure 2. Different methods used to generate diversity using the S. cerevisiae toolbox. (A) epPCR from a single template followed by in vivo recombination in S. cerevisiae. (B) In vivo DNA shuffling. Several parental genes are recombined and cloned with a linearized vector into S. cerevisiae in a single step. (C) epPCR in conjunction with in vivo DNA shuffling. (D) IvAM (In vivo Assembly of Mutant libraries with different mutational spectra). Two or more distinct mutant libraries are generated by epPCR using polymerases with different biases. S. cerevisiae is transformed with the mutant libraries together with the linearized plasmid. (E) IVOE for combinatorial saturation mutagenesis or site-directed mutagenesis. The gene is amplified in two independent PCR reactions using mutagenized/degenerate primers. By engineering specific overhangs, the PCR products are then cloned into S. cerevisiae together with the linearized plasmid in a single transformation. (F) Mutagenic StEP (Staggered Extension Process). Several parental genes are used as templates during mutagenic StEP, promoting the random introduction of mutations during the short cycles of annealing and extension. The resulting mutant/recombined library is further shuffled by S. cerevisiae, together with the linearized plasmid. Stars represent single mutations.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.