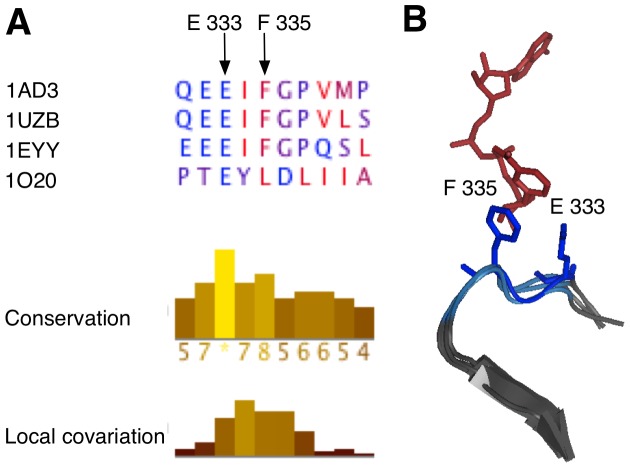
Figure 5. Local covariation identifies active site residues.
(A) Screenshot from the LoCo tool showing the region of high local covariation from BAliBASE 3 alignment BB11003. BB11003 is an alignment of four paralogous oxireductases with similar structure. The local covariation peaks highlight four positions in the sequence alignment which are coloured in blue in panel B. Two active site residues from structure 1AD3, E333 and F335, are emphasized in the sequence alignment. (B) Structure alignment of residues shown in panel A made in PyMOL [40]. The region of high local covariation is highlighted in blue; structure 1AD3 is emphasized with dark blue. The NAD cofactor from structure 1AD3 is drawn in red. Important binding residues E333 and F335 from 1AD3 are rendered in sticks representation.