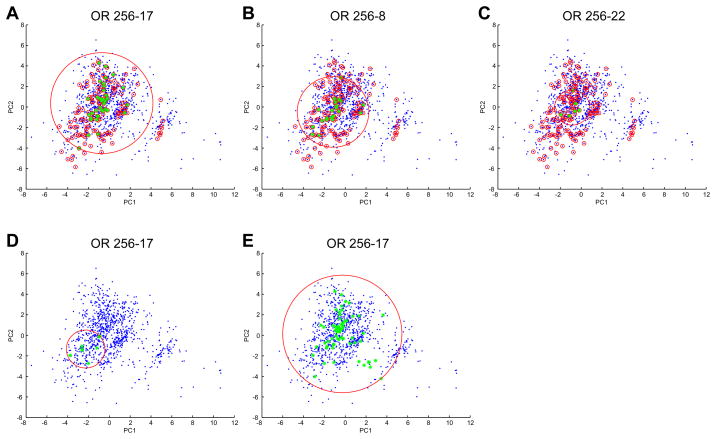
Figure 7. MOR256-17 is broadly tuned.
Odor space was estimated using 1035 odorants (blue dots) in a multi-dimensional odor space was based on 32 optimized physicochemical descriptors (Haddad et al., 2008) and first and second principal components were used to plot this odor space. A 32D hypersphere enclosing all 1035 odorants in this odor space has a radius of 18.5. The 155 odorants in our screening set are indicated by small red circles. The large red circle in each panel is a 2D representation of a 32D hypersphere that encloses the odorants that activate the receptor. A) Odorants from the screening set that activate MOR256-17 are indicated by green stars. A 32D hypersphere that encloses the odorants that activate MOR256-17 has a radius of 11.4. B) Odorants from the screening set that activate MOR256-8 are indicated by green stars. A 32D hypersphere that encloses these odorants has a radius of 5.3. C) Odorants from the screening set that activate MOR256-22 are indicated by green stars. A 32D hypersphere that encloses these odorants has a radius of 4. D) Odorants previously shown to activate MOR256-17 (Dahoun et al., 2011) are indicated by green stars. A 32D hypersphere that encloses these odorants has a radius of 4.8. E) All odorants shown to activate MOR256-17 in this study are indicated by green stars. A 32D hypersphere that encloses these odorants has a radius of 16.4.