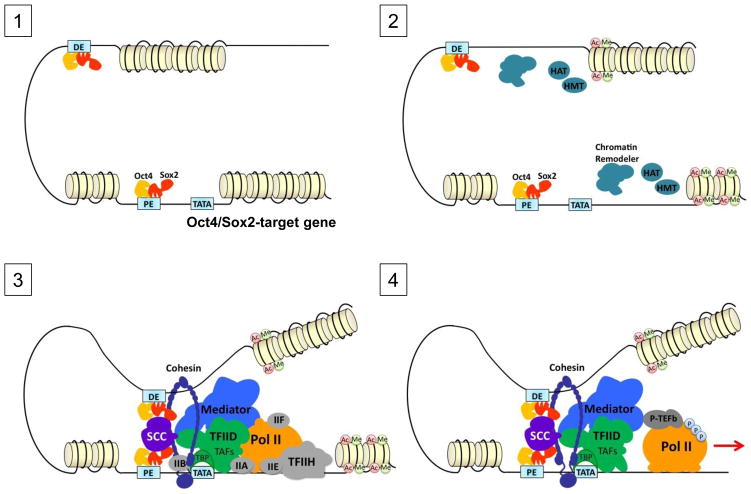
Figure 1.
Multi-step activation model of an Oct4/Sox2-target gene in embryonic stem cells. The transcription apparatus is assembled on an Oct4/Sox2-target gene promoter in a spatially and temporally regulated manner to initiate gene expression. The stepwise assembly is depicted here in the order proposed in [68]. (1) ‘Orchestration’: sequence-specific transcription factors, Oct4 and Sox2, bind to both the proximal (PE) and distal (DE) enhancers. (2) ‘Access’: Chromatin remodelers and histone modifying enzymes such as histone acetyltransferases (HATs) and histone methyltransferases (HMTs) act coordinately to remodel chromatin around the gene loci by altering nucleosome positioning and histone modifications. This allows access of other transcription factors and cofactors to the gene promoter. (3) ‘Initiation’: Preinitiation complex (PIC) and various coactivators (e.g. TAFs/TFIID, Mediator and SCC) are assembled onto the core promoter via interaction with activators bound on PE and DE (e.g. TAFs-Oct4/Sox2 or SCC-Oct4/Sox2), promoter DNA elements (e.g. TBP-TATA), and modified nucleosomes (e.g. TAF3-H3K4me3). DNA looping by cohesin and Mediator can further stabilize the long range enhancer-promoter DNA interaction. (4) ‘Elongation’: phosphorylation of the C-terminal domain (CTD) of the largest subunit of RNA polymerase II (Pol II) by TFIIH facilitates promoter escape while subsequent phosphorylation by P-TEFb stimulates the transition of Pol II into productive elongation.