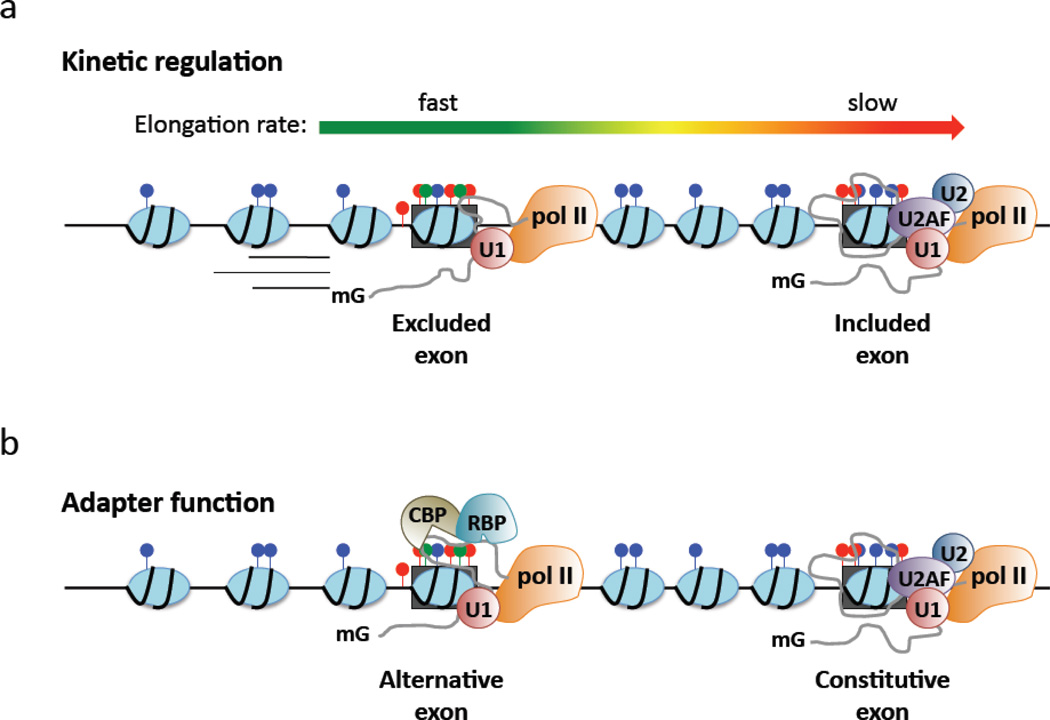
Figure 2. Potential mechanisms for chromatin-associated spliceosome assembly.
Exons are marked by increased nucleosome occupancy, distinct histone modifications and elevated DNA methylation relative to introns. These modifications at the DNA level may influence splice site selection by a) modulating elongation or b) through direct recruitment of auxiliary factors. a) A slow rate of pol II elongation favors spliceosome assembly at weak exons, whereas a rapid rate may not provide a sufficient spatiotemporal window prior to synthesis of competing downstream splice sites. Intragenic chromatin structure may act to locally modulate elongation rate. As shown here, the upstream exon is excluded from mRNA as a result of a locally rapid pol II elongation rate, thereby shifting spliceosome assembly to the downstream exon. b) Chromatin modifications may recruit chromatin binding proteins (CBP) to exonic DNA, which thereby act as adaptor molecules for RNA binding proteins (RBP) that promote or inhibit spliceosome assembly. As shown here, the chromatin context of the upstream exon binds a CBP that recruits an RNA binding splicing repressor, thereby shifting spliceosome assembly to the downstream exon.