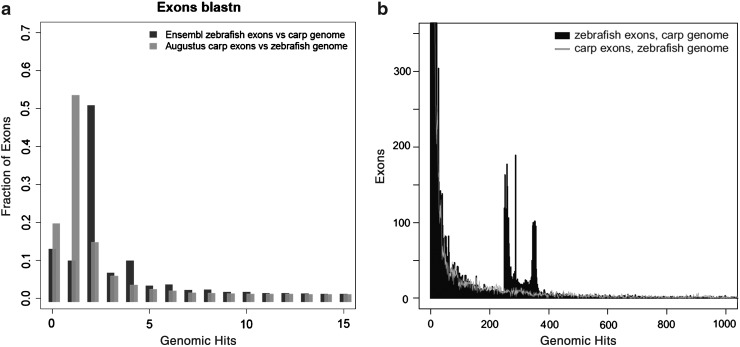
FIG. 5.
Quantitation of the multiplicity of BLAST hits. (a) BLASTN of predicted carp exons versus the zebrafish genome results in a single best hit in the majority of cases (gray bars). In contrast, BLASTN of ENSEMBL zebrafish exons versus the carp genome often yields two hits (black). In both cases, an E-value cutoff of 1e–10 was used. Similar results are obtained using zebrafish exons predicted by Augustus instead of annotated by ENSEMBL (Supplementary Fig. S3). In Supplementary Figure S4, we performed a control BLASTN analysis of zebrafish exons versus the zebrafish genome confirming that the results show that zebrafish is different to common carp in its number of highly repetitive exon predictions. (b) The same histogram as (a) but at larger scale. Although most zebrafish exons show a single hit on the carp genome, a subset matches a very large number of loci in carp. In contrast, the distribution of hits of carp exons on the zebrafish genome is more even.