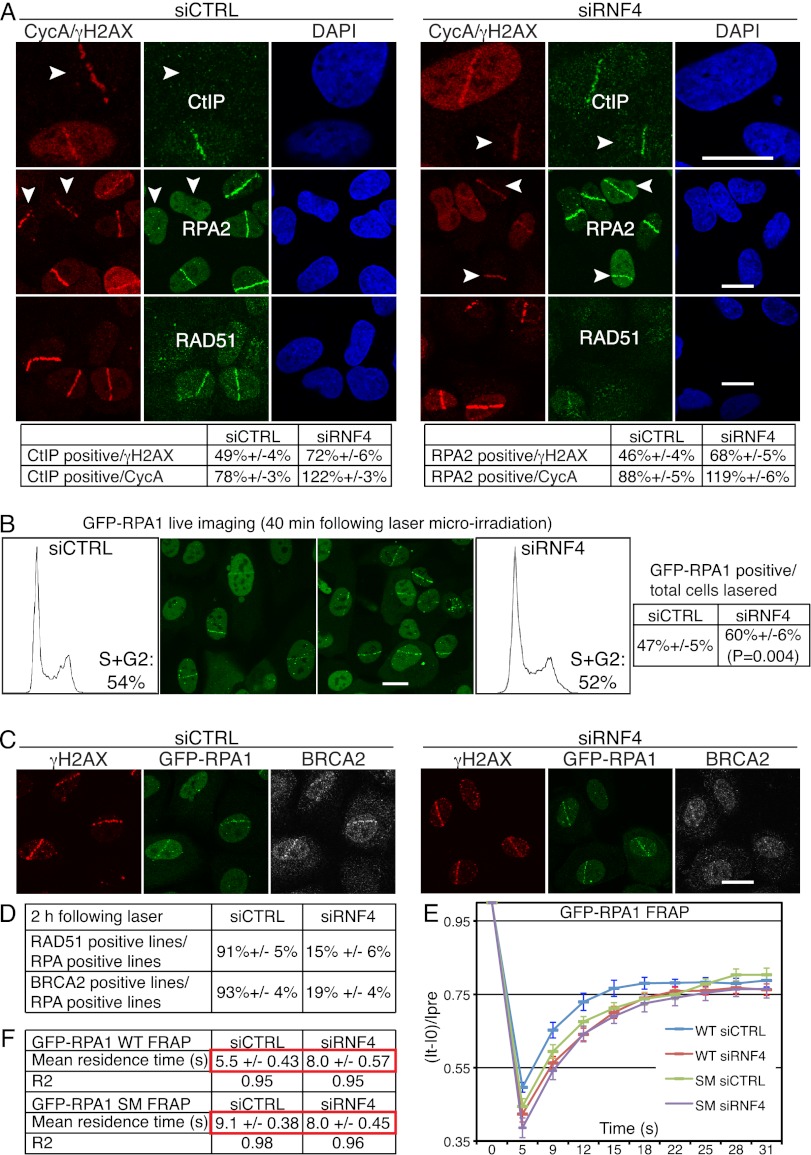
Figure 4.
RNF4 regulates RPA turnover and BRCA2-mediated RAD51 loading onto damaged DNA. (A) RNF4 is required for normal accumulation of CtIP and RPA2 and for efficient accumulation of RAD51 at DNA damage sites. U2OS cells transfected with siRNAs were laser-micro-irradiated, fixed after 2 h, and then analyzed by immunofluorescence as indicated; arrows mark cyclinA (CycA)-negative cells. For additional images of RPA staining and cyclinA immunoblotting in control and RNF4-depleted cells, see Supplemental Figure S5A. Quantification numbers represent proportions of cells showing CtIP or RPA2 accumulation out of γH2AX- or cyclinA-positive cells ±SED (n > 100). (Note that the staining of RPA2 and CtIP in cyclinA-negative cells is unlikely to be left over from the previous G2, as these cells would have to have been lasered/damaged in S/G2 and would then have had to progress through mitosis with high amounts of damage even though they possess an intact G2/M checkpoint [see Fig. 3F], all within the 2-h incubation step of this experiment.) (B) U2OS cells stably expressing GFP-RPA1 (wild-type [WT]) were transfected with siRNAs and laser-micro-irradiated 48 h later (to allow subsequent quantification, the numbers of cells lasered were documented). GFP-RPA1 lines were live-imaged and counted 40 min post-irradiation. Proportions of cells displaying GFP-RPA1 lines out of total cells irradiated are presented ±SED with the corresponding flow cytometry profiles (n > 150 cells per siRNA, accumulated over three independent experiments). (C) RNF4 is required of efficient accumulation of BRCA2 at DNA-damaged sites. U2OS cells stably expressing GFP-RPA1 were treated as in A and stained with the indicated antibodies. For additional images of RAD51 and BRCA2 accumulation in GFP-RPA1 control and RNF4-depleted cells, see Supplemental Figure S5, B and C. (D) Quantifications of RAD51- and BRCA2-positive cells in GFP-RPA1-positive control and RNF4-deleted cells (n ≥ 100 cells per siRNA, ±SED). (E,F) U2OS cells stably expressing GFP-RPA1 wild-type (WT) or GFP-RPA1K449R,K577R (SM) were transfected with siRNAs and, 48 h later, laser-micro-irradiated and subjected to FRAP analysis 40 min post-irradiation (n = 24 independent measurements; error bars, ±SED). For equations and calculations, see the Materials and Methods. For siRNA depletions, see Supplemental Figure S11A.