Summary
Autosomal Recessive Primary Microcephaly (MCPH) is characterized by small brain size due to deficient neuron production in the developing cerebral cortex. While MCPH is a rare disease, the questions surrounding its etiology strike at the core of stem cell biology. The seven genes implicated in MCPH all encode centrosomal proteins and disruption of the MCPH gene Cdk5rap2 in mice revealed its role in neural progenitor proliferation and in maintaining normal centriole replication control. Here, we discuss the impact that centrosome regulation has on neural progenitors in the developing brain. We integrate the impact of centriole replication defects with the functions of Cdk5rap2 and other MCPH proteins, propose mechanisms for progenitor loss in MCPH, and discuss links to two other microcephaly syndromes.
MCPH: A centrosome-based disease
The cerebral cortex is the region of the brain where learning, memory, language and other cognitive and motor activities are controlled. Although the relative and actual size of the cerebral cortex varies among mammals, it develops in a similar way [1]. The mouse is therefore an appropriate model to investigate human cerebral cortex development.
Neurogenesis of the cerebral cortex requires coordinated temporal control of neural precursor cell divisions during embryonic brain development. In the mammalian brain, neural progenitors reside adjacent to the lateral ventricles in the ventricular and subventricular zones. Neuronal patterning of the cerebral cortex occurs by radial migration of new neurons outward from the ventricular and subventricular zones of the neocortex [1]. The six layers formed during corticogenesis are produced in an “inside-out” manner. The first neurons populate the innermost layer, while those produced later migrate in the radial direction past the early neurons to occupy the outermost layers. The ventricular zone is a pseudostratified columnar epithelial layer surrounding the ventricle and is where early symmetric divisions expand the apical progenitor pool. Then, between embryonic days 10-12 in the mouse, the polarity of progenitor divisions changes, becoming progressively more asymmetric. Asymmetric progenitor divisions are neurogenic, producing a daughter neuron or neuronal precursor and a renewed stem cell [2, 3]. Basal progenitors, produced from asymmetric division of apical progenitors, undergo a single neurogenic division [4]. There are systematic changes in the proportion of symmetric vs. asymmetric apical progenitor divisions during neocortical development [5], and control over this process is critical for the accurate and appropriate production of neurons at the cerebral cortex [6].
Mutations in the MCPH genes reduce the population of neurons in each of the six layers of the brain during development, yet with little or no overt structural abnormalities other than reduced thickness of the cerebral cortex [7, 8]. MCPH individuals generally have reduced cognitive function but no motor deficit. MCPH loci map to at least seven genes, MCPH 1-7 [7-16] (Table 1).
TABLE 1.
The MCPH genes
| MCPH gene |
OMIM 251200 |
Map location |
Other names | Paralog† |
Drosophila ortholog |
Centrosome or spindle pole |
DNA damage response |
|---|---|---|---|---|---|---|---|
| MCPH1 |
251200 607117 |
8p23 | Microcephalin, BRIT1 |
None | MCPH1 | Yes | Yes |
| MCPH2 |
604317 613583 |
19q13.12 | WDR62 | MAPKBP1 | CG7337 | Yes | ND |
| MCPH3 |
604804 608201 |
9q33.3 | Cdk5rap2, Cep215 |
Myomegalin/ PDE4DIP |
Centrosomin (Cnn) |
Yes | Yes |
| MCPH4 |
604321 613529 |
15q21.1 | Cep152 |
C10orf118/ CTCL tumor antigen L14-2 |
Asterless (Asl) |
Yes | Yes |
| MCPH5 |
608716 605481 |
1q31 | Aspm, Calmbp1 |
hSfi1/ Sfi1 |
Abnormal spindle (Asp) |
Yes | Yes |
| MCPH6 |
608393 609279 |
13q12.2 | CPAP, CENPJ, Sas-4 |
TCP10 | Sas-4 | Yes | ND |
| MCPH7 |
612703 181590 |
1p32 | STIL, SIL, Sas-5 |
None? | Ana2 | Yes | ND |
Except for Myomegalin/PDE4DIP and MAPKBP1, the paralogs listed here are proposed based upon similarity searches by the authors. ND: not determined.
Surprisingly, all the defined MCPH genes encode centrosome or spindle pole proteins (Table 1), implicating proper centrosome or mitotic microtubule organization as critical to generating and/or maintaining normal neuron populations in the developing brain. In recent years, mutations in an expanding number of genes involved in centriole and cilium assembly and function have been linked to a spectrum of ciliopathies [17-19]. The revelation that many diseases are based on defects in cilia and centrosomes has highlighted the importance of understanding the cellular and developmental functions of centrosomes.
The cell biological cause for MCPH, whether it be aberrant cilia, mitotic centrosomes, or other centrosomal basis has not been demonstrated. Moreover, several MCPH proteins function in the DNA damage response pathway involving ATR (ATM and Rad3-related). Thus, there are two common threads emerging in MCPH disease: the centrosome and DNA damage response. How they connect is not clear. Recently, however, several reports indicate that Cdk5rap2 is required for the maintenance of apical neural progenitors in the mouse brain, for a normal DNA damage response, and to restrict centriole duplication [20-23]. Loss of the latter activity results in multipolar spindles, and in cells with more than one primary cilium. This commentary will focus on Cdk5rap2 and how recent findings, in combination with findings on other MCPH proteins, reveal important clues into the cell biology of MCPH as a centrosome-based disease.
The centrosome
The centrosome is the primary microtubule-organizing center (MTOC) in animal cells. It is a large complex consisting of a pair of centrioles surrounded by a pericentriolar matrix (PCM) and is present in nearly all animal cells (Box1). The centrosome regulates many cellular processes including cell polarity and cell division, in addition to functioning as a hub for factors required for cell cycle progression and DNA damage response [24, 25].
Box 1. The Centrosome.
A typical cell contains one or two centrosomes, depending on the cell cycle stage. Each centrosome contains a pair of centrioles. The centriole is a microtubule-based cylindrical structure with nine-fold radial symmetry [24] (Figure I). A nascent “daughter” centriole assembles at the proximal end of the “mother” centriole and remains tightly engaged to the mother centriole until anaphase, when the mother-daughter bond is disengaged in a process that requires Plk1 and separase [80], yet the pair remain linked via a fibrous linker consisting of rootletin, C-Nap1 and Cep68, and regulated by Nek1 kinase, protein phosphatase 1, Cdk5rap2 and other proteins ([74] and references therein) (Figure I).
In contrast to mutations in their mammalian counterparts, mutations in some MCPH ortholog genes in Drosophila (Table 1) result in complete loss of centrioles. Centrioles are absent in Sas-4 [73] or asterless (asl) [81] loss-of-function mutant flies. Moreover, Asl/CEP152 serves an essential early role in centriole biogenesis [56, 57, 81, 82]. In contrast to Drosophila, where development to adults can be achieved without centrioles, mouse development arrests early if centrioles are severely disrupted. This is because centrioles are required for assembly of cilia, which are essential for hedgehog and other signaling pathways [17, 18]. Yet, in humans, mutations in the asl and Sas-4 orthologs CEP152 and CPAP result in MCPH or SCKL. Moreover, loss of asp in Drosophila causes mitotic arrest and lethality, while severe loss-of-function mutations in ASPM are viable [11]. In addition, in Drosophila, mutants for the single CDK5RAP2 ortholog cnn fail to assemble PCM and mitotic MTOCs are severely deficient [83], a very different phenotype from Cdk5rap2 mutant cells which have apparently normal, albeit amplified, mitotic centrosomes [20]. Since MCPH proteins are conserved and likely share functional attributes, there are probably paralogs in human that supply functional redundancy, but for which neural progenitors are uniquely sensitive [11] (Table 1).
Paralogs for CDK5RAP2 and WDR62, have been reported (MYOMEGALIN and MAPKBP1, respectively). Table 1 shows the results of paralog searches conducted using iterative BLAST (PSI BLAST), revealing the presence of potential paralogs for CEP152, ASPM and CPAP (Table 1). Therefore, tissue-specific or paralog-specific functions may be ascribed to Cdk5rap2 and other MCPH genes in neural progenitors [11]. A thorough survey of the expression patterns of these genes will be necessary to determine if paralog redundancy limits the effects of MCPH mutations.
The diverse cellular functions of the centrosome portend a range of mechanisms that affect the pathology of MCPH, such as progenitor mitotic spindle defects or other cell cycle disruptions, interkinetic nuclear migration in apical progenitors, cilium dysfunction, or neuronal migration. Since cortical patterning of neurons appears relatively normal in MCPH brains, neuronal migration is an unlikely etiology. To understand which aspects of centrosome function are impacted in MCPH it is essential to study the roles of the MCPH proteins using in vivo models. Recent studies on Cdk5rap2 and other MCPH genes have provided important clues [20-23, 26-28].
Cdk5rap2 function at the centrosome
Cdk5rap2 is a centrosomin family protein. Centrosomin orthologs have been extensively characterized in the fruit fly Drosophila melanogaster and the fission yeast S. pombe, where these proteins are essential for MTOC activity (Box 2). RNAi knockdown and mutation of Cdk5rap2 reveals diverse centrosomal functions and its requirement for neural progenitor division or survival (Table 2). From these investigations, a variety of activities are attributed to Cdk5rap2 including centrosome cohesion, centriole engagement, centriole replication control, MTOC activity, gamma tubulin recruitment, attachment of centrosomes to spindle poles, binding to the dynactin complex, DNA damage response, Chk1 kinase recruitment, microtubule plus end dynamics, checkpoint control, and spindle orientation in neural progenitors. Linking these cellular phenotypes to function in the developing brain using vertebrate model systems is critical to understand the mechanisms of cerebral cortex development and the basis of MCPH.
Box 2. Functions of centrosomin family proteins.
Centrosomin family members are conserved in eukaryotes, yet appear absent from plants. The founding member, centrosomin (CNN), was discovered in Drosophila melanogaster. Centrosomin proteins have extensive coiled-coil domains, and two highly conserved domains, Centrosomin Motifs 1 and 2 (CM1 and CM2) (Figure II). The functions for centrosomins are most extensively characterized for the Drosophila and the yeast S. pombe orthologs, CNN and Mto1, respectively. Null mutations in cnn disrupt assembly of the pericentriolar matrix (PCM) surrounding the centrioles in the early embryonic centrosomes, and at mitosis in somatic cells [83]. A similar loss of MTOC activity occurs in S. pombe mto1 mutants ([84] and references therein). Centrioles detach from spindle poles in cnn mutants [30, 85], similar to what occurs in Cdk5rap2 mutant chicken cells [23]. The CM1 domain is required for centrosome separation in Drosophila embryos, and for efficient recruitment of γ-tubulin, TACC and Msps [30]. In chicken DT40 cells, Cdk5rap2 CM1 was required to recruit AKAP450 and the p150Glued subunit of dynactin to centrosomes [23]. In S. pombe, CM1 is necessary but not sufficient to recruit γ-TuC to MTOCs [86]. Human CDK5RAP2 CM1, on the other hand, was reported to bind directly to γ-TuRC and activate it [87]. Mto1 acts cooperatively with Mto2, which has no obvious orthologs in flies or vertebrates, to recruit γ-tubulin [86]. Since vertebrates have a clear paralog to Cdk5rap2 in myomegalin, it will be interesting to see if depletion of both causes loss of centrosomal MTOC activity as occurs with mutation of the single ortholog in Drosophila and S. pombe.
TABLE 2.
Cdk5rap2 depletion phenotypes
| Method of depletion |
System | Knockdown or mutant phenotypes and other activities | Reference |
|---|---|---|---|
| Mutation | Human | Microcephaly. Centrosomal defects not determined. | [10] |
|
RNAi |
U2OS, A549 and hTERT- RPE1 Cell culture |
Loss of cohesion. Pcnt was required for Cdk5rap2 recruitment. |
[74] |
| RNAi | HeLa, U2OS and MRC-5 |
Loss of MTOC activity, and failure to recruit γ-TuRC to centrosomes. CM1 domain binds to γ-TuRC. |
[75] |
| RNAi | HeLa | Mitotic spindle checkpoint defective. Cdk5rap2 associates with Cdc20. Binds to checkpoint gene promoters in nuclei and regulates their expression. |
[76] |
| RNAi | HeLa | Modest reduction in γ-tubulin recruitment to mitotic centrosomes. Co-dependence of Cdk5rap2 and pericentrin for centrosome localization. Plk1 required for recruitment of Cdk5rap2 to centrosomes. |
[77] |
| RNAi | U2OS | Cdk5rap2 binds to microtubule plus ends and is dependent on EB1, a direct binding partner. Activity is not conserved in rodents. |
[78] |
| RNAi | HeLa | Cdk5rap2 recruits dynein to centrosomes and depends on dynactin for its recruitment to centrosomes. |
[79] |
|
Mutation / RNAi |
Targeted disruption in chicken DT40 cells/ HeLa |
CM1 and CM2 domains each required for attachment of centrosomes to spindle poles. Cdk5rap2 recruits AKAP450 and p150glued/dynactin to centrosomes. Subtle reduction of γ-tubulin at Cdk5rap2 CM1 mutant centrosomes. Defective DNA damage response. Reduced Chk1 kinase recruitment to centrosomes. Loss of cohesion (HeLa, RNAi). |
[23] |
|
RNAi in utero |
Mouse embryonic brain |
Premature neuronal differentiation. Reduced proliferation and increased cell cycle exit by neural progenitors. Loss of apical but increase in basal progenitors. Cdk5rap2 and pericentrin interact. Pericentrin recruits Cdk5rap2 to centrosomes, but not vice-versa. |
[21] |
| Originally identified as Hertwig’s anemia (an), with hematopoietic progenitor proliferation deficiency, elevated aneuploidy, radiation sensitivity, lack of primordial germ cells, and early onset histiocytic sarcomas. Phenotypes are strain dependent. |
[29]. |
||
|
Mutation |
Mouse |
Mutation (an) mapped to an inversion of exon 4 that deletes most of CM1. Severe microcephaly, but is strain dependent. Cortical neurons are most deficient in later- forming superficial layers. Reduction in neural progenitors, especially basal progenitors at later stages of neurogenesis. Apparent mitotic delay. Multipolar spindles in neuronal precursors. Defective spindle orientation in cortical progenitors. Mitotic centrosomes recruit Aurora A kinase. Premature cell cycle exit and increased cell death of neuronal progenitors. |
[22] |
| Mutation | Mouse | Two gene trap mutations (Cdk5rap2RRU031 and Cdk5rap2RRF465) that produce truncated proteins. Centriole amplification in embryonic fibroblasts. Loss of centriole cohesion and engagement. Multipolar spindles and supernumerary cilia. Mitotic delay. Mitotic centrosomes recruit γ-tubulin and other PCM components and are apparently normal MTOCs. No microcephaly observed. |
[20] |
HeLa, U2OS, A549, hTERT-RPE1 and MRC-5 are human cell lines.
CM1 and CM2 are centrosomin motifs 1 and 2, respectively.
Mouse models for MCPH
Three mouse mutants for Cdk5rap2 were reported recently. One mutation, originally associated with Hertwig’s anemia (an) mutant mouse, is a gamma radiation-induced mutation that inverts exon 4 [22, 29]. The inverted exon is spliced out of the transcript, resulting in a 37 amino acid deletion that removes most of CM1 (Box 2, Figure I). Homozygous Cdk5rap2an mice have microcephaly. The small cerebral cortex correlates with multipolar spindles, misalignment of spindles in neural precursors, and loss of neural progenitors [22]. Cdk5rap2an mice show loss of primordial germ cells and infertility, anemia with hematopoietic precursor proliferation defects, aneuploidy, increased generation of histiocytic sarcoma, and radiation sensitivity. While rescue with a Cdk5rap2 transgene would confirm the linkage of these phenotypes, linkage is likely because the an mutation has been backcrossed over dozens of generations and the an allele failed to complement another Cdk5rap2 mutant (Cdk5rap2RRO242)[22, 29]. Surprisingly, microcephaly was strain-dependent, as were the other Cdk5rap2an phenotypes [22, 29]. The existence of such background effects in humans is an intriguing possibility.
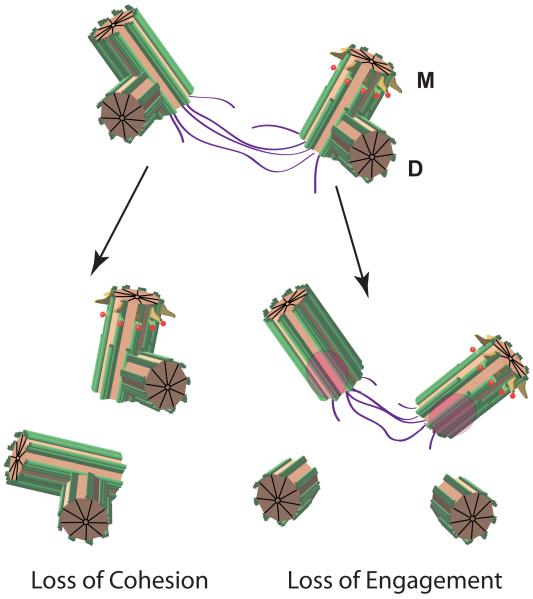
Figure I. Forms of centrosomal bondage.
Centriole mother-daughter pairs are tightly bound in the “engaged” state until their release is triggered, which normally occurs at mitosis. The spot vacated by the procentriole is shown in pink, and is a trigger to license replication. The older mother (M) is marked with distal (yellow) and subdistal (red) appendages. The daughter (D) is shown as a small procentriole. A linker (purple) comprised of the related proteins C-Nap1 and Rootletin connects each centriole pair. The linker connects the mother centrioles between centrosomes. Loss of cohesion and engagement normally occur in the cell cycle at G2 and anaphase, respectively. In homozygous Cdk5rap2RRF465 mutant cells, cohesion and engagement are both disrupted.
Human MCPH is not associated with anemia or early onset cancers, possibly reflecting differences between the Cdk5rap2an mouse model and human disease. Notably, there are no reports on the fertility of MCPH individuals for comparison to the Cdk5rap2an mouse. A possible explanation for the phenotype differences between human and Cdk5rap2an mouse could be the types of mutations. The Cdk5rap2an mouse has an in-frame deletion affecting CM1, yet the mutant protein is expressed and localized at centrosomes. In contrast, the human alleles are conceptual protein truncations (Box2, Figure I) [10]. In Drosophila, a deletion mutation within the CM1 domain of cnn, similar to Cdk5rap2an, causes centrosome separation failure in embryos, a dominant and neomorphic phenotype [30]. The Cdk5rap2an mutation was not dominant, yet a neomorphic function cannot be excluded. In contrast to the mouse Cdk5rap2an mutation, a similar CM1 deletion mutation in chicken DT40 cells did not cause centrosome amplification or multipolar spindles, but rather centrosome disjunction from the spindle and defective DNA damage response ([23] and Table 2).
In another study, two slice trap mutations, resulting in 64 and 435 amino acid truncations, designated Cdk5rap2RRU031 and Cdk5rap2RRF465 respectively, were characterized in mice [20]. These alleles are similar to the two mapped human CDK5RAP2 mutations (Box 2, Figure I) [10]. Neither mutant, however, displayed microcephaly or other obvious defects in brain development, although background effect, as seen for the Cdk5rap2an mouse, was not tested. Of the two mutations, one (Cdk5rap2RRU031) was leaky, producing about 7% expression of full-length protein relative to wild type in cultured embryonic fibroblasts (MEFs). Despite low expression, the protein was detected at centrosomes. On the other hand, Cdk5rap2RRF465 appears fully penetrant, expressing only the truncated protein. Both mutants were defective in centrosome cohesion coincident with reduced centrosomal signal for rootletin (Box 1), yet Cdk5rap2RRF465 also resulted in centriole amplification. Loss of centriole engagement (Box 1) was the evident cause of amplification in Cdk5rap2RRF465 mutant MEFs, but de novo centriole biogenesis [31] was not excluded as an alternative mechanism. The consequences of centriole amplification in Cdk5rap2RRF465 mutant MEFs are multipolar spindles and supernumerary primary cilia. How these phenotypes may contribute to MCPH is discussed below. Comparison of Cdk5rap2 mutant phenotypes to other mouse MCPH mutants will be important to establish whether common or disparate mechanisms account for MCPH.
In addition to Cdk5rap2, mouse mutants for Aspm and Mcph1 were constructed. Two mouse mutants, one short (Aspm1-7) and one long (Aspm1-25) truncation of Aspm, produced modest microcephaly (Aspm1-7 had a neocortex that was 86% the thickness of control) [26]. Embryonic Aspm mutant brains showed no significant differences in the symmetry of progenitor divisions. In contrast, Aspm knockdown by RNAi in utero increased the ratio of asymmetric to symmetric divisions of neural progenitors, an effect that would conceptually curtail expansion of the apical progenitor pool in an in vivo context [32]. The Aspm mutants also showed reduced numbers of germ cells and reduced male and female fertility. Interestingly, human Aspm rescued microcephaly in the mouse mutant (to a normal mouse-sized cerebral cortex) [26].
Two Mcph1 mutants, one a targeted knockout of exon 2 (Mcph1−) [28], the other a splice trap insertion (Mcph1gt) [27] were generated. The Mcph1gt mutant displayed no microcephaly or DNA damage sensitivity. The Mcph1gt mutation had leaky expression through the splice trap insertion (28% mRNA expression relative to wild type in the brain), yet still displayed the premature chromosome condensation phenotype associated with MCPH1 mutation in humans [7, 8, 11]. The Mcph1− mutant, on the other hand, had reduced birth rate, slow growth, radiation sensitivity, defective DNA damage response, and male infertility due to meiotic arrest and apoptosis of spermatocytes. Brain size was not examined in the Mcph1− mouse. Centrosome aberrations were not reported in the Aspm or Mcph1 mouse mutants. Thus, despite the generation of several MCPH mouse models, a clear etiology for MCPH has not emerged.
What common functions emerge from MCPH models?
Given that MCPH proteins reside at centrosome or spindle poles, a plausible basis for MCPH lies with centrosome function and/or spindle assembly. On the other hand, defective response to DNA damage is another common feature. Whether these two features are related is unclear. Examination of centrosomal and DNA damage response phenotypes among all animal models with microcephaly will be important to determine if these are universal MCPH phenotypes.
Centrosome amplification occurs in Cdk5rap2an and Cdk5rap2RRF465 mice, in MCPH1 mutant human and chicken cells [33, 34], and may also occur in CEP152 mutant human cells [35]. This shared phenotype suggests that centrosome amplification may be a common link in MCPH. Centrosome amplification can disrupt the fidelity and timing of mitosis, spindle orientation during stem cell division (Box 3), and be tumorigenic [19, 36-38]. In Cdk5rap2an and Cdk5rap2RRF465 mutants, amplified centrosomes assemble multipolar spindles in MEFs [20] and neural progenitors [22], with the latter result correlated to progenitor loss in the Cdk5rap2an embryonic brain. In addition, excess centrioles lead to supernumerary primary cilia in affected cells [20].
Box 3. Regulation of neural stem cell division polarity.
A fundamental tenet of stem cell biology is the control of polarized division during organogenesis to produce two cells destined with different fates. One cell differentiates, while the other becomes a renewed stem cell that continues the division process to populate differentiated tissues. Spindle orientation during neural progenitor cell divisions requires both cortical determinants that capture astral microtubules, and functional centrosomes that generate the astral microtubules (see Figure 1). Spindle orientation becomes random in neural progenitors when centrosomes are inactivated. In Drosophila, determinants that polarize neuroblasts and the centrosomes that respond to polarization cues are essential for normal stem cell proliferation and for the coordinated asymmetric division that leads to production of neural precursors and stem cell self-renewal [6, 41, 42]. Disruption of polarization or spindle orientation cues can result in increased symmetric divisions and metastatic tumorigenesis [88]. A similar paradigm functions in mouse neural progenitors, except that polarization cues and spindle orientation appear to actively control symmetric rather than asymmetric divisions. The loss of which may result in increased neurogenic asymmetric divisions [6, 41, 42]. Disruption of LGN, a conserved neural stem cell polarizing component, however, caused randomized spindle orientation in mouse and chick [46, 47], yet had no impact on brain size in mice [46]. Thus, the importance of spindle orientation in progenitor cells is called into question [6, 41, 42, 46, 47, 89].
Centrosome asymmetry, on the other hand, may be a conserved feature of stem cell biology. In Drosophila, male germline stem cells maintain the older “mother” centrosome near the apical cell membrane proximal to the niche, a microenvironment that maintains stem cells [90]. Similarly, in neuroblasts one centrosome is maintained near the apical membrane and has MTOC activity, while the other is inactive until late in prophase when it migrates to the basal side of the cell [43, 44]. In contrast to germ cells, the apically-anchored centrosome is the younger centrosome in Drosophila neuroblasts [50]. Recent studies [52] show that, opposite to Drosophila neuroblasts, mouse neural progenitors maintain the older mother centriole when they divide asymmetrically in the mouse brain, and may rely on centrosome asymmetry to maintain progenitor pools. Therefore, the principle of centrosome asymmetry may be a common regulatory mechanism for polarized stem cell division.
While posing a threat to chromosomal stability, multipolar spindles activate the mitotic checkpoint, allowing time for extra centrosomes to cluster and for the spindle to resolve into a bipolar structure [38]. Nevertheless, aneuploidy can result due to inappropriate microtubule-chromosome attachments, leading in most cases to cell death [37]. Whether chromosomal instability, or other cell cycle effects, is responsible for MCPH in Cdk5rap2 mutants is unclear. However, in the Cdk5rap2an mouse, neural progenitor cells were progressively lost, coincident with premature cell cycle exit and elevated apoptosis [22]. Similarly, in utero RNAi of Cdk5rap2 in embryonic brains caused premature cell cycle exit and loss of apical progenitors, with a resulting increase of basal progenitors and differentiated neurons during the 3-day time-course of the experiments [21]. Notably, Cdk5rap2RRF465 MEFs and Cdk5rap2 mutant chicken cells had premature senescence [20, 23]. Therefore cell cycle disruption is a possible factor in MCPH, a role implicated for Mcph1 [11, 27, 33, 39]. Consistent with this scenario, when cell cycle exit is manipulated directly using p27Kip1 overexpression, neocortical cell numbers are reduced similarly to MCPH [40].
Spindle orientation
Another possible effect of centrosome amplification is an impact on mitotic spindle orientation in neural progenitors. Spindle polarity may influence the control and timing of cell fate determination [6, 41, 42] (Box 3). Spindle orientation becomes random in neural progenitors when centrosomes are inactivated, or upon disruption of microtubule asters that link the spindle to the cell cortex, or disruption of the Lis1/dynactin complex that links asters to the cell cortex to regulate spindle rotation [42]. Supernumerary centrosomes, induced by Plk4 overexpression, lead to spindle misorientation in Drosophila [36]. In normal Drosophila neuroblasts the apical centrosome is a “dominant” MTOC, with a larger PCM (Figure 1) [43, 44]. This centrosome asymmetry was lost among amplified centrosomes in Plk4-overexpressing neuroblasts. Moreover, neuroblasts overproliferated, presumably due to increased symmetric, rather than asymmetric, divisions [36]. An opposite effect, a shift from symmetric to asymmetric division, might be expected with mammalian progenitors under similar conditions (Figure 1,Box 3).
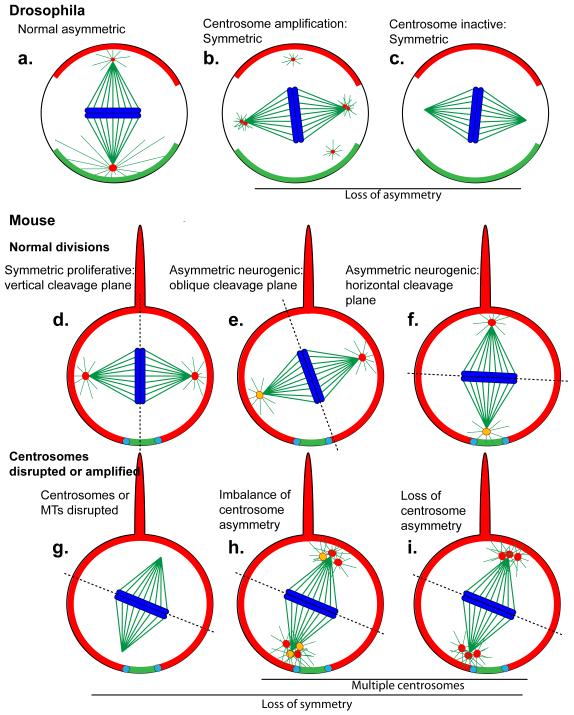
Figure 1. Models for control of progenitor division symmetry by centrosomes.
Centrosomes assemble astral microtubules that link the spindle to polarizing determinants localized at the cell cortex. Astral microtubules thereby link spindle orientation to cell polarity. (a) In Drosophila neuroblasts the centrosomes are asymmetric, where the younger “daughter” centrosome assembles a larger PCM and a larger aster while the older “mother” centrosome undergoes PCM loss in prophase, followed by PCM restoration at prometaphase, and then the older centrosome segregates into the ganglion mother cell [43, 44, 50, 72]. (b) When centrosomes are amplified, this asymmetry is lost and neuroblasts divide symmetrically [36]. (c) When astral microtubules or the dynactin complex are disrupted, the spindle is misoriented early in mitosis but gets corrected towards telophase [41, 42]. Neuroblasts frequently divide symmetrically, however, when centrosomes are completely absent [19, 41, 42, 73].
Neural progenitor cells in mouse divide symmetrically during the proliferative phase (d), and then switch to an asymmetric mode of division upon neurogenesis (e,f). If the spindle is misaligned during the proliferative divisions the cell may divide asymmetrically (e,f) [3, 6, 32, 41, 45]. The centrosomes are asymmetrically inherited, with the older mother centriole (yellow centrosome) retained in the apical progenitor cell (the opposite to Drosophila, where the younger centriole is retained in the progenitor cell) [52].
How amplified centrosomes or rearrangement of mother centrioles will impact progenitor division in mice is not known. (g) Symmetric division is disrupted if centrosomes or the dynactin complex are disrupted. The drawings in (h) and (i) are speculative impacts that centrosome amplification may have. In (h), imbalance of positioning of mother centrioles could impact spindle rotation, while in (i), centrosome asymmetry may be lost, similar to what was observed in Drosophila (b).
In (a-i), the basal cortical determinants are labeled red, and the apical green. The centrosomes are red, the microtubules green, and the chromosomes blue. In (d-i), the projection on top of the cells is the basal process, shown here not to scale (it is relatively much longer). The dotted line delineates the cleavage furrow, the adherens junctions are indicated in light blue.
During neurogenesis of the mouse cerebral cortex, neural progenitors initially divide symmetrically to expand the progenitor pool, and then switch to an asymmetric phase for neurogenesis. Since the apical cortical aspect is small in apical epithelial progenitors (Figure 1), even a slight skew in the cleavage plane angle can promote asymmetric inheritance of cortical determinants [6]. Mutation of Cdk5rap2 or Nde1 (a Lis1/dynactin complex component) or RNAi of Aspm alters spindle orientation in apical progenitors. These phenotypes correlated with progenitor depletion (and reduced cerebral cortex in the case of Nde1 and Cdk5rap2) [22, 32, 45]. One interpretation of these results is that precocious asymmetric division produces neurons early at the expense of progenitor pool expansion. In this model, an active centrosome-dependent process drives the symmetric proliferative divisions, the loss of which results in asymmetric division. In conflict with this idea, however, depletion of LGN, a non-centrosomal polarity determinant, caused randomized spindle orientation and apical progenitors were displaced from the mouse neuroepithelium. Nevertheless, this did not affect progenitor pools, which relocated basally, or cerebral cortex development [46, 47]. Moreover, disruption of aPKCλ in mouse brain disrupts apical adherens junctions, interkinetic nuclear migration, and the neuroepithelial tissue architecture [48]. Despite these disruptions, cortical neurogenesis is not impeded. Therefore, while centrosome disruption may impact spindle orientation in mouse neural progenitor cells, it is clear that spindle misorientation is not sufficient to disrupt neurogenesis and cause MCPH.
Centrosome asymmetry
While the centrosome pair can regulate spindle orientation in polarized cells, they are also asymmetric, one being older than the other. Centrosome asymmetry was first described in Drosophila male germline stem cells where the older “mother” maintains association with the apical cortex [49]. In Drosophila neuroblasts, on the other hand, the younger “daughter” centrosome is maintained apically and the older “mother” is inherited by the neuronal precursor cells [50]. This relationship may not be important, however, because asymmetric division occurs successfully when the centrosomes are experimentally manipulated into switched positions [51]. In the embryonic mouse brain, the older mother centriole is retained in the renewed apical progenitor, while the younger mother is distributed into differentiated cells [52]. Depletion of ninein, a mother centriole-specific marker, by in utero RNAi disrupted this asymmetry and neural progenitors were depleted [52]. Together, these data suggest that centrosome asymmetry may be an important regulator of neurogenesis, somehow impacting progenitor fate or survival. Imbalance of the mother and daughter centriole numbers or ratios could, under this scenario, disrupt progenitor division polarity and survival in mouse Cdk5rap2 mutant neural progenitors (Figure 1).
The centrosome in neural progenitor maintenance: a role for cilia
Centriole disruption or amplification could impact neural progenitor survival by means other than mitotic spindle or cell cycle disruption. Primary cilia are key signaling centers for hedgehog (hh) and other signaling pathways [17, 18], and have established roles in neural progenitor maintenance at the neocortex [53]. Most human cells contain a single primary cilium, whose assembly is templated by the mother centriole. Consistent with a cilium basis for MCPH, the Stil mutant mouse displayed defects in Sonic hh signaling, resulting in neural tube and left-right patterning defects consistent with cilium dysfunction [7, 8].
MEFs from Cdk5rap2RRF465 mice exhibited multiple primary cilia in cells with amplified centrioles. Whether this is also true for the neural progenitor cells, particularly in the microcephalic Cdk5rap2an mutant, was not investigated. Supernumerary cilia could create a quantitative change in intercellular signaling, thereby impacting neural progenitors. In support of this model, a recent study showed that, in cells with multiple primary cilia induced by Plk4 overexpression, that hh signal reception was reduced by approximately 50% in cells with two primary cilia compared to cells with one (Tim Stearns, personal communication). Thus, supernumerary cilia may dampen rather than amplify hh signaling, and this could impact survival of progenitor cells. Regulation of cilium assembly and function is therefore a possibility for all the MCPH proteins, given the connection they share to the centrosome. Among the MCPH proteins, Cdk5rap2, Cep152, CPAP or their orthologs have been shown to impact cilium assembly or function in some form. It is noteworthy that pericentrin (see below) and ninein are also required for cilium assembly [54, 55].
If indeed the basis for MCPH is centrosomal, it remains to be shown whether the cause is disruption of cilia, spindle regulation at mitosis, or some other yet unknown basis. Given the diverse functions attributed thus far to the seven known MCPH proteins, multiple causes are a possibility [7, 8]. Recently, however, several of the MCPH orthologs (Table 1) were found to reside in complexes in Drosophila (Cnn, Asl, and Sas-4) and human cells (CEP152 and CPAP) [56-58]. With MCPH proteins assembled together, it is plausible they function through a common pathway to impact brain development. In addition, the centrosomal protein pericentrin (Pcnt) [54] binds directly to Cdk5rap2 [21, 59], and associates with several MCPH proteins or their Drosophila orthologs (Box 4) [39, 58].
Box 4. Cdk5rap2 and Pericentrin are partners.
Pericentrin (Pcnt) is a conserved centrosomal protein mutated in MOPD II [54]. Evidence from studies in fungi, Drosophila and human cells point to roles for Pcnt and its orthologs in regulation of microtubule assembly into the mitotic spindle. It is also required for cilium assembly, which appears to be its critical function in Drosophila [54]. Interactions between Cdk5rap2 and PCNT were shown for the human, mouse and Drosophila homologs ([21, 58, 59] and our unpublished data). The Cdk5rap2-Pcnt association was shown to be direct for the mouse and human counterparts. How Pcnt and Cdk5rap2 are functionally related is unclear, but RNAi knockdown studies in mouse neural progenitors and in cell lines showed that Pcnt is required for Cdk5rap2 localization to centrosomes, while Cdk5rap2 is not required for Pcnt localization [21, 59, 74], except in HeLa cells where they are mutually dependent [74, 77]. Pcnt RNAi affected neural progenitor proliferation similarly to Cdk5rap2 knockdown [21]. PCNT mutant cells did not show amplified centrosomes, but they did show a reduction or absence of the microtubule nucleator γ-tubulin at centrosomes, as did mutation of the gene for MCPH1, which also associates with PCNT [39, 60]. PCNT and MCPH1 cooperate to recruit CHK1 kinase to centrosomes to regulate mitotic entry. While Pcnt is not reported to affect centrosome replication, disruption of its partner MCPH1 causes centriole amplification in some cell types [34]. Tying these connections together will be important to understand the functional significance of the CDK5RAP2-PCNT interaction. Moreover, the intimate association between CDK5RAP2 and PCNT suggests a shared etiology between MCPH and MOPD II.
MCPH has links to SCKL and MOPD II syndromes
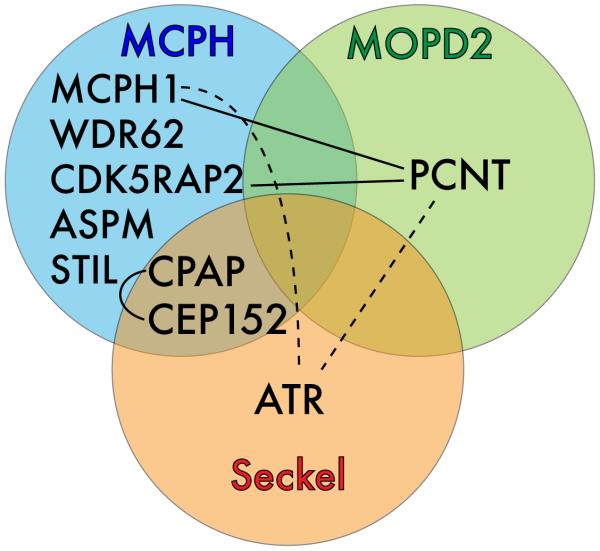
PCNT is mutated in microcephalic osteodysplastic primordial dwarfism Majewski type II (MOPD II, MIM 210720) [60, 61]. Primordial dwarfism (proportionately small overall stature) and microcephaly are among the clinical features of MOPD II. These features overlap with Seckel syndrome (SCKL1, MIM 210600), and some patients with lesions in PCNT were diagnosed as SCKL. A consensus is emerging, however, that mutations in PCNT are assigned only to MOPD II [62, 63]. Recently, CPAP and CEP152 mutations, originally mapped in MCPH, were also mapped to SCKL syndrome [35, 64]. Other MCPH mutations also cause short stature [7, 12], yet this is a prominent feature in SCKL and MOPD II. Interestingly, there appears no correlation between the perceived severity of the lesions mapped to CPAP and its association with MCPH versus SCKL, and may reflect genetic background effects [64]. The similarities of MCPH, SCKL and MOPD II syndromes have been noted frequently [7, 8, 54, 64], and the recent genetic and molecular findings underscore relatedness among these microcephaly syndromes (Figure 2). Among these syndromes, there is a common thread to DNA damage response.
Figure 2. Convergence of three centrosome-based microcephaly diseases.
A Venn diagram showing that MCPH, MOPD II and SCKL Syndromes have mutations in common genes, and exhibit physical interactions (connecting lines) and indirect interactions (via CHK1, the effector kinase for ATR; dashed lines) that connect these diseases into a group of with possible common centrosomal etiology. Not shown are additional interactions shown in Drosophila, that link the orthologs of CDK5RAP2, CPAP, CEP152 and PCNT in a common complex.
Cell cycle arrest in response to DNA damage is a critical checkpoint at S-phase or G2/M to ensure repair of insults prior to cell cycle resumption. These responses require the ATM (ataxia telangiectasia, mutated) and ATR kinase signaling cascades acting through the effector checkpoint kinases Chk1 and Chk2 [65]. Chk1 and Chk2 localize to centrosomes, where Chk1 inhibits cyclin-dependent kinase 1 (Cdk1) activation and halts the cell cycle at the G2/M transition in response to DNA damage [65].
Centrosome amplification is a hallmark response to DNA damage or to aberrant DNA damage signaling [65]. Amplification of centrosomes in response to ionizing radiation is Chk1-dependent [25, 66]. SCKL cells, whether mutant in ATR or other genes, exhibit centrosome amplification [67]. It will be informative to survey cells from CPAP and CEP152 SCKL patients, and from MCPH patients, for amplified centrosomes and impaired DNA damage response to determine if either or both of these effects are shared in these syndromes.
Mcph1, Cdk5rap2, Cep152 and Aspm, like PCNT and ATR [39, 60], have roles in, or responses to, the DNA damage response [7, 8, 23, 33, 35, 39, 68, 69]. MCPH1 coordinates with PCNT to recruit CHK1 to centrosomes where it inhibits CDC25B [39]. Mcph1 acts upstream of ATR but downstream of γ-H2AX in foci formation on ionizing radiation-induced DNA damage in chicken cells [68]. Importantly, MCPH1 mutant or knockdown cells show centrosome amplification, depending on the cell type, with hyper-amplification upon DNA damage [33, 34]. Moreover, DNA damage-induced centrosome duplication appears to involve centriole disengagement [70], similar to the proposed mechanism for centriole amplification in Cdk5rap2RRF465 mouse cells [20].
The centrosome amplification and DNA damage response impairment observed in Seckel, MOPD II, and MCPH cells, and the multiple interactions found among the proteins involved, link these syndromes at a cellular and molecular level. These links notwithstanding, it is unclear what causal connection exists between the DNA damage response and microcephaly diseases. Does impaired DNA damage response have a direct impact on neural progenitors, or is the effect an indirect one due to centriole amplification or other centrosome disruption? On the other hand, the possibility that amplification or other centrosome effects is an inconsequential side effect of impaired DNA damage signaling in MCPH cannot be excluded.
While the MCPH mutations produce small brains, the MOPD II and SCKL mutations also produce small overall stature, suggesting that while MCPH affects primarily neural progenitors, the MOPD II and SCKL mutations affect the stem cells for multiple organs. In support of this, a conditional knockout for ATR causes progeria in adult mice and a progressive loss of stem cells in multiple tissues [71]. Since ATR mutant SCKL cells have amplified centrosomes [33], reminiscent of Cdk5rap2 mutant mouse cells [20, 22], perhaps a related mechanism depletes stem cells in both mutants and between syndromes.
Future Directions
Deciphering the functions of CDK5RAP2 and the other MCPH proteins will reveal fundamental aspects of neural stem cell biology. The findings that mutations in Cdk5rap2 mutant mice produce amplified centrosomes and neural progenitor loss supports previous models whereby MCPH arises from a centrosome-based defect in neural stem cell division or maintenance. Drawing on recent findings in a variety of systems regarding MCPH protein functions and the roles of centrosomes in neural stem cells, several models are invoked to explain how MCPH may disrupt neural progenitor cells. The distinguishing features of these prevailing models involve centrosome roles in i) mitotic spindle orientation, ii) centrosome asymmetry iii) cilium assembly or function, and iv) DNA damage response. In the neocortex one or more of these likely contribute to neuron reduction in MCPH by affecting cell proliferation, specifically cell cycle exit, or possibly also cell survival. While excess centrosomes are a feature of Cdk5rap2, MCPH1, and CEP152 mutant cells, it remains to be seen if this is common among MCPH mutations, or if some other common feature unites the etiology. Exciting discoveries are on the horizon as the mechanisms that link the MCPH proteins to the biology of neural stem cells are exposed. Experimental linkage of the molecular deficits to the specific changes in the behavior of the neural stem cells that produce the neocortex presents an exciting challenge for MCPH and also for MOPD II and SCKL. The full impact of centrosome disruption in Cdk5rap2 mutant cells in vivo remains to be understood, but it is clear that the regulatory roles of centrosomes are crucial for the mechanisms of stem cell control in organogenesis.
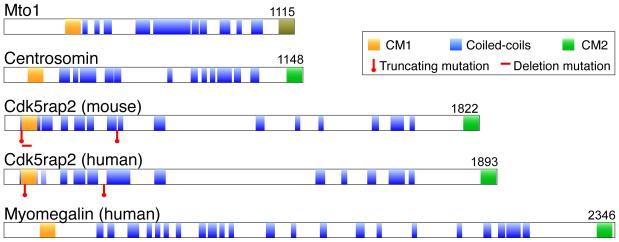
Figure II. Cdk5rap2 is a centrosomin family member.
Schematic diagram shows the relative domain structures of centrosomin proteins from S. pombe (Mto1), Drosophila (Centrosomin), mouse (Cdk5rap2), and both human paralogs (Cdk5rap2 and Myomegalin). Mice and other vertebrates also have Myomegalin (not shown). The relative positions of conserved domains are shown, and positions of mutations are indicated for mouse and human Cdk5rap2. The C-terminal box in Mto1 (shaded in olive green) is not conserved with CM2 in the metazoans shown, but it is required for localization to MTOCs, similar to the function of CM2.
Acknowledgements
We thank Ling-Rong Kao and Tomer Avidor-Reiss for comments and discussion, and funding support from NIH grant R01GM068756, and an ARRA supplement to support GM068756, from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 3.Buchman JJ, Tsai LH. Spindle regulation in neural precursors of flies and mammals. Nat Rev Neurosci. 2007;8:89–100. doi: 10.1038/nrn2058. [DOI] [PubMed] [Google Scholar]
- 4.Noctor SC, et al. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J Comp Neurol. 2008;508:28–44. doi: 10.1002/cne.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caviness VS, et al. Histogenetic processes leading to the laminated neocortex: migration is only a part of the story. Dev Neurosci. 2008;30:82–95. doi: 10.1159/000109854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong W, Chia W. Neurogenesis and asymmetric cell division. Curr Opin Neurobiol. 2008;18:4–11. doi: 10.1016/j.conb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Kaindl AM, et al. Many roads lead to primary autosomal recessive microcephaly. Prog Neurobiol. 2010;90:363–383. doi: 10.1016/j.pneurobio.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Thornton GK, Woods CG. Primary microcephaly: do all roads lead to Rome? Trends in Genetics. 2009;25:501–510. doi: 10.1016/j.tig.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bilguvar K, et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bond J, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- 11.Bond J, Woods CG. Cytoskeletal genes regulating brain size. Curr Opin Cell Biol. 2006;18:95–101. doi: 10.1016/j.ceb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Darvish H, et al. A clinical and molecular genetic study of 112 Iranian families with primary microcephaly. J Med Genet. 2010;47:823–828. doi: 10.1136/jmg.2009.076398. [DOI] [PubMed] [Google Scholar]
- 13.Guernsey DL, et al. Mutations in centrosomal protein CEP152 in primary microcephaly families linked to MCPH4. Am J Hum Genet. 2010;87:40–51. doi: 10.1016/j.ajhg.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar A, et al. Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am J Hum Genet. 2009;84:286–290. doi: 10.1016/j.ajhg.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholas AK, et al. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat Genet. 2010 doi: 10.1038/ng.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu TW, et al. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat Genet. 2010 doi: 10.1038/ng.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berbari NF, et al. The primary cilium as a complex signaling center. Curr Biol. 2009;19:R526–535. doi: 10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 20.Barrera JA, et al. CDK5RAP2 regulates centriole engagement and cohesion in mice. Dev Cell. 2010;18:913–926. doi: 10.1016/j.devcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchman JJ, et al. Cdk5rap2 Interacts with Pericentrin to Maintain the Neural Progenitor Pool in the Developing Neocortex. Neuron. 2010;66:386–402. doi: 10.1016/j.neuron.2010.03.036. [DOI] [PubMed] [Google Scholar]
- 22.Lizarraga SB, et al. Cdk5rap2 regulates centrosome function and chromosome segregation in neuronal progenitors. Development. 2010;137:1907–1917. doi: 10.1242/dev.040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barr AR, et al. CDK5RAP2 functions in centrosome to spindle pole attachment and DNA damage response. The Journal of Cell Biology. 2010;189:23–39. doi: 10.1083/jcb.200912163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- 25.Shimada M, Komatsu K. Emerging connection between centrosome and DNA repair machinery. J Radiat Res (Tokyo) 2009;50:295–301. doi: 10.1269/jrr.09039. [DOI] [PubMed] [Google Scholar]
- 26.Pulvers JN, et al. Mutations in mouse Aspm (abnormal spindle-like microcephaly associated) cause not only microcephaly but also major defects in the germline. Proc Natl Acad Sci U S A. 2010;107:16595–16600. doi: 10.1073/pnas.1010494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trimborn M, et al. Establishment of a mouse model with misregulated chromosome condensation due to defective Mcph1 function. PLoS One. 2010;5:e9242. doi: 10.1371/journal.pone.0009242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang Y, et al. BRIT1/MCPH1 is essential for mitotic and meiotic recombination DNA repair and maintaining genomic stability in mice. PLoS Genet. 2010;6:e1000826. doi: 10.1371/journal.pgen.1000826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barker JE, et al. High incidence, early onset of histiocytic sarcomas in mice with Hertwig’s anemia. Exp Hematol. 2005;33:1118–1129. doi: 10.1016/j.exphem.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Megraw TL. Proper recruitment of gamma-tubulin and D-TACC/Msps to embryonic Drosophila centrosomes requires Centrosomin Motif 1. Mol Biol Cell. 2007;18:4037–4049. doi: 10.1091/mbc.E07-05-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loncarek J, Khodjakov A. Ab ovo or de novo? Mechanisms of centriole duplication. Mol Cells. 2009;27:135–142. doi: 10.1007/s10059-009-0017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fish JL, et al. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci U S A. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alderton GK, et al. Regulation of mitotic entry by microcephalin and its overlap with ATR signalling. Nat Cell Biol. 2006;8:725–733. doi: 10.1038/ncb1431. [DOI] [PubMed] [Google Scholar]
- 34.Brown JA, et al. MCPH1/BRIT1 limits ionizing radiation-induced centrosome amplification. Oncogene. 2010;29:5537–5544. doi: 10.1038/onc.2010.302. [DOI] [PubMed] [Google Scholar]
- 35.Kalay E, et al. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat Genet. 2010 doi: 10.1038/ng.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basto R, et al. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ganem NJ, et al. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Acilan C, Saunders WS. A tale of too many centrosomes. Cell. 2008;134:572–575. doi: 10.1016/j.cell.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 39.Tibelius A, et al. Microcephalin and pericentrin regulate mitotic entry via centrosome-associated Chk1. J Cell Biol. 2009;185:1149–1157. doi: 10.1083/jcb.200810159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tarui T, et al. Overexpression of p27 Kip 1, probability of cell cycle exit, and laminar destination of neocortical neurons. Cereb Cortex. 2005;15:1343–1355. doi: 10.1093/cercor/bhi017. [DOI] [PubMed] [Google Scholar]
- 41.Lesage B, et al. Neural stem cells: the need for a proper orientation. Curr Opin Genet Dev. 2010;20:438–442. doi: 10.1016/j.gde.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 42.Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- 43.Rebollo E, et al. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev Cell. 2007;12:467–474. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 44.Rusan NM, Peifer M. A role for a novel centrosome cycle in asymmetric cell division. J Cell Biol. 2007;177:13–20. doi: 10.1083/jcb.200612140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng Y, Walsh CA. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 46.Konno D, et al. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat Cell Biol. 2008;10:93–101. doi: 10.1038/ncb1673. [DOI] [PubMed] [Google Scholar]
- 47.Morin X, et al. Control of planar divisions by the G-protein regulator LGN maintains progenitors in the chick neuroepithelium. Nat Neurosci. 2007;10:1440–1448. doi: 10.1038/nn1984. [DOI] [PubMed] [Google Scholar]
- 48.Imai F, et al. Inactivation of aPKClambda results in the loss of adherens junctions in neuroepithelial cells without affecting neurogenesis in mouse neocortex. Development. 2006;133:1735–1744. doi: 10.1242/dev.02330. [DOI] [PubMed] [Google Scholar]
- 49.Yamashita YM, Fuller MT. Asymmetric centrosome behavior and the mechanisms of stem cell division. J Cell Biol. 2008;180:261–266. doi: 10.1083/jcb.200707083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conduit PT, Raff JW. Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in drosophila neuroblasts. Curr Biol. 2010;20:2187–2192. doi: 10.1016/j.cub.2010.11.055. [DOI] [PubMed] [Google Scholar]
- 51.Januschke J, Gonzalez C. The interphase microtubule aster is a determinant of asymmetric division orientation in Drosophila neuroblasts. J Cell Biol. 2010;188:693–706. doi: 10.1083/jcb.200905024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, et al. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han YG, Alvarez-Buylla A. Role of primary cilia in brain development and cancer. Curr Opin Neurobiol. 2010;20:58–67. doi: 10.1016/j.conb.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Delaval B, Doxsey SJ. Pericentrin in cellular function and disease. The Journal of Cell Biology. 2010;188:181–190. doi: 10.1083/jcb.200908114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Graser S, et al. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cizmecioglu O, et al. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J Cell Biol. 2010;191:731–739. doi: 10.1083/jcb.201007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dzhindzhev NS, et al. Asterless is a scaffold for the onset of centriole assembly. Nature. 2010 doi: 10.1038/nature09445. [DOI] [PubMed] [Google Scholar]
- 58.Conduit PT, et al. Centrioles regulate centrosome size by controlling the rate of Cnn incorporation into the PCM. Curr Biol. 2010;20:2178–2186. doi: 10.1016/j.cub.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 59.Wang Z, et al. Conserved motif of CDK5RAP2 mediates its localization to centrosomes and the Golgi complex. J Biol Chem. 2010;285:22658–22665. doi: 10.1074/jbc.M110.105965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Griffith E, et al. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat Genet. 2008;40:232–236. doi: 10.1038/ng.2007.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rauch A, et al. Mutations in the Pericentrin (PCNT) Gene Cause Primordial Dwarfism. Science. 2008;319:816–819. doi: 10.1126/science.1151174. [DOI] [PubMed] [Google Scholar]
- 62.Willems M, et al. Molecular analysis of Pericentrin gene (PCNT) in a series of 24 Seckel/MOPD II families. J Med Genet. 2009 doi: 10.1136/jmg.2009.067298. [DOI] [PubMed] [Google Scholar]
- 63.Piane M, et al. Majewski osteodysplastic primordial dwarfism type II (MOPD II) syndrome previously diagnosed as Seckel syndrome: report of a novel mutation of the PCNT gene. Am J Med Genet A. 2009;149A:2452–2456. doi: 10.1002/ajmg.a.33035. [DOI] [PubMed] [Google Scholar]
- 64.Al-Dosari MS, et al. Novel CENPJ mutation causes Seckel syndrome. J Med Genet. 2010;47:411–414. doi: 10.1136/jmg.2009.076646. [DOI] [PubMed] [Google Scholar]
- 65.Loffler H, et al. Structure meets function--centrosomes, genome maintenance and the DNA damage response. Exp Cell Res. 2006;312:2633–2640. doi: 10.1016/j.yexcr.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 66.Bourke E, et al. DNA damage induces Chk1-dependent centrosome amplification. EMBO Rep. 2007;8:603–609. doi: 10.1038/sj.embor.7400962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alderton GK, et al. Seckel syndrome exhibits cellular features demonstrating defects in the ATR-signalling pathway. Hum Mol Genet. 2004;13:3127–3138. doi: 10.1093/hmg/ddh335. [DOI] [PubMed] [Google Scholar]
- 68.Jeffers LJ, et al. Distinct BRCT domains in Mcph1/Brit1 mediate ionizing radiation-induced focus formation and centrosomal localization. Oncogene. 2008;27:139–144. doi: 10.1038/sj.onc.1210595. [DOI] [PubMed] [Google Scholar]
- 69.Fujimori A, et al. Ionizing radiation downregulates ASPM, a gene responsible for microcephaly in humans. Biochem Biophys Res Commun. 2008;369:953–957. doi: 10.1016/j.bbrc.2008.02.149. [DOI] [PubMed] [Google Scholar]
- 70.Inanc B, et al. A Centrosome-autonomous Signal that Involves Centriole Disengagement Permits Centrosome Duplication in G2 Phase After DNA Damage. Mol Biol Cell. 2010 doi: 10.1091/mbc.E10-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ruzankina Y, et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Januschke J, et al. Drosophila neuroblasts retain the daughter centrosome. Nat Commun. 2011;2:243. doi: 10.1038/ncomms1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Basto R, et al. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 74.Graser S, et al. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J Cell Sci. 2007;120:4321–4331. doi: 10.1242/jcs.020248. [DOI] [PubMed] [Google Scholar]
- 75.Fong KW, et al. CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol Biol Cell. 2008;19:115–125. doi: 10.1091/mbc.E07-04-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang X, et al. CDK5RAP2 is required for spindle checkpoint function. Cell Cycle. 2009;8:1206–1216. doi: 10.4161/cc.8.8.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haren L, et al. Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS One. 2009;4:e5976. doi: 10.1371/journal.pone.0005976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fong KW, et al. Interaction of CDK5RAP2 with EB1 to track growing microtubule tips and to regulate microtubule dynamics. Mol Biol Cell. 2009;20:3660–3670. doi: 10.1091/mbc.E09-01-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee S, Rhee K. CEP215 is involved in the dynein-dependent accumulation of pericentriolar matrix proteins for spindle pole formation. Cell Cycle. 2010;9:774–783. [PubMed] [Google Scholar]
- 80.Tsou MF, et al. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell. 2009;17:344–354. doi: 10.1016/j.devcel.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Blachon S, et al. Drosophila asterless and vertebrate Cep152 Are orthologs essential for centriole duplication. Genetics. 2008;180:2081–2094. doi: 10.1534/genetics.108.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hatch EM, et al. Cep152 interacts with Plk4 and is required for centriole duplication. J Cell Biol. 2010;191:721–729. doi: 10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Megraw TL, et al. Zygotic development without functional mitotic centrosomes. Curr Biol. 2001;11:116–120. doi: 10.1016/s0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- 84.Samejima I, et al. Fission yeast Mto1 regulates diversity of cytoplasmic microtubule organizing centers. Curr Biol. 2010;20:1959–1965. doi: 10.1016/j.cub.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lucas EP, Raff JW. Maintaining the proper connection between the centrioles and the pericentriolar matrix requires Drosophila centrosomin. J Cell Biol. 2007;178:725–732. doi: 10.1083/jcb.200704081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Samejima I, et al. Two distinct regions of Mto1 are required for normal microtubule nucleation and efficient association with the gamma-tubulin complex in vivo. J Cell Sci. 2008;121:3971–3980. doi: 10.1242/jcs.038414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Choi YK, et al. CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J Cell Biol. 2010;191:1089–1095. doi: 10.1083/jcb.201007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gonzalez C. Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat Rev Genet. 2007;8:462–472. doi: 10.1038/nrg2103. [DOI] [PubMed] [Google Scholar]
- 89.Farkas LM, Huttner WB. The cell biology of neural stem and progenitor cells and its significance for their proliferation versus differentiation during mammalian brain development. Curr Opin Cell Biol. 2008;20:707–715. doi: 10.1016/j.ceb.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 90.Yamashita YM, et al. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]