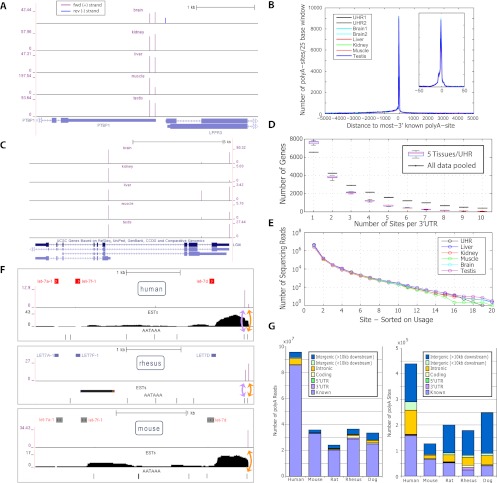
Figure 3.
Assessment of basic features of the PolyA-seq atlas. (A) PolyA-seq detects polyA sites in a strand-specific manner. Two polyA sites (vertical spikes) are detected in human splicing factor PTBP1 (forward genomic strand, indicated by arrows) in all tissues, while LPPR3 (reverse strand) has a single polyA site, detected only in brain. Y-axis units are reads per million (see Methods; note that y-axis scales vary among tissues). PolyA-seq sites on the forward and reverse genomic strands are shown in different colors. (B) Human sites agree to single-base precision with known transcript termini. Known termini represent the 3′-most site reported by RefSeq, UCSC KG, or Ensembl per gene. (C) PolyA-seq reveals constitutive and tissue-dependent polyA sites. In human LGI4, polyA site choice is governed by alternative splicing. The 5′-most site is used in all tissues even as absolute expression levels fluctuate (see A for details). The intermediate site is used primarily in liver, while the downstream site is repressed in kidney, but is otherwise expressed at levels similar to the upstream site. (D) Number of polyA sites/3′UTR in five human tissues and UHR (navg/tissue = 16,387, ntotal uniq = 20,873; see Methods for 3′UTR compilation). All samples were normalized to equal numbers of aligned sequencing reads by random selection. (Black lines) Sites/3′UTR for aggregated data from these six samples. (E) Number of sequencing reads/site; sites were selected based on decreasing order of usage per 3′UTR. (F) Lineage-dependent polyadenylation of a pri-microRNA transcript. PolyA-seq detects polyadenylation downstream from the microRNA cluster containing let7a1, let7f1, and let7d in all tissues assayed in all species (data not shown, but see Supplemental Fig. 8 for additional details; for simplicity, PolyA-seq data and polyA signals are shown here only for human, rhesus, and mouse kidney, and only for the sense strand; arrows within microRNA precursors indicate the direction of transcription). In human and rhesus, two polyA sites (purple spikes) correspond to two canonical polyA signals (AATAAA; black tick marks), the first of which is present only in primate genomes (data not shown). In rat, mouse, and dog, only the downstream polyA site is detected, in accordance with the absence of the upstream polyA signal. (G) Distribution of reads and polyA sites across genomic features. All reads were aggregated in each species and then filtered and clustered as described in the main text.