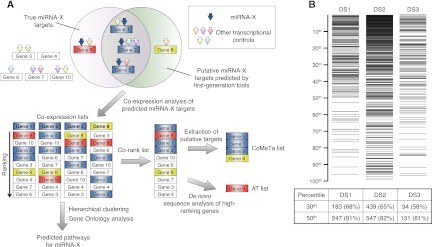
Figure 1.
The CoMeTa procedure. (A) For simplicity, the strategy is described on a subgroup of 10 genes. Multiple transcriptional controls (arrows) for these genes are shown, including a specific miRNA (miRNA-X, blue arrows). In the example, Gene 8 (yellow box) and Gene 9 (red box) are, respectively, a false positive and a false negative result of sequence-based software predictions, used to seed the analysis. In co-expression lists, the ranking of the genes is an index of their expression correlations with the probe gene. A co-rank list is obtained by averaging the co-expression lists. The false-positive target ranks low in the co-rank list, whereas the false-negative target ranks high and can be identified by subsequent de novo sequence analyses (additional targets [AT] list). The CoMeTa output consists of the list of predicted targets ranked by expression analysis. Co-expression lists are subsequently used for further analysis (see text). (B) Distribution of known miRNA targets (horizontal lines in the top panel) within CoMeTa lists, for three independent data sets of miRNA targets (DS1, DS2, DS3). Counts within the first 30th and 50th percentiles are provided for each data set in the lower table. The average number of targets for each miRNA present in the first 50th percentile of CoMeTa lists is 750 (DS1), 850 (DS2), and 900 (DS3). P-values: <10−39 (DS1), <10−29 (DS2), and <10−23 (DS3).