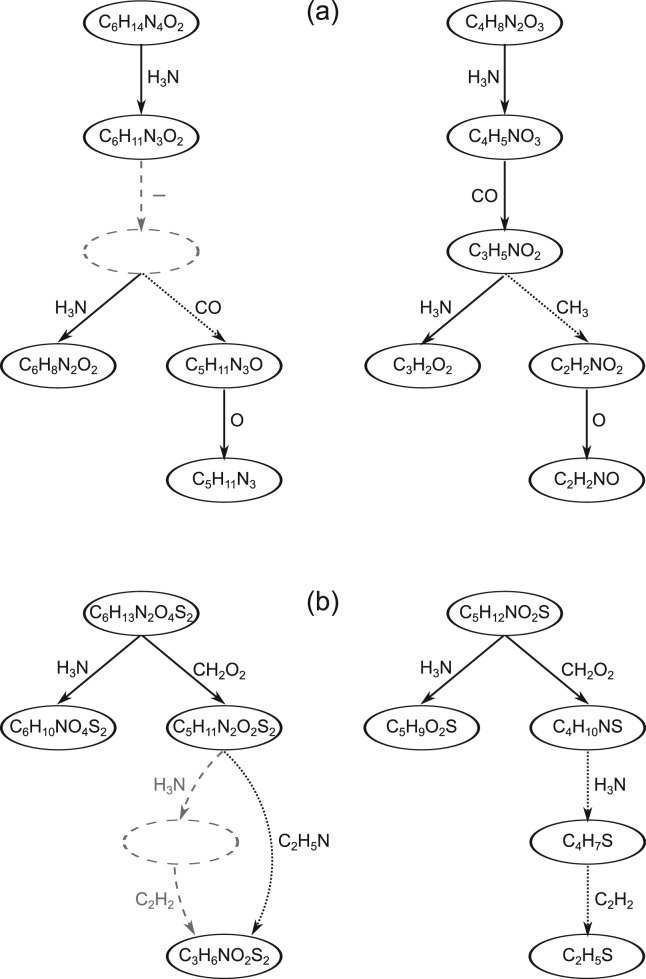
Fig. 2.
Two alignments of fragmentation trees based on edge similarities. Nodes represent molecular formulas of the fragments, edges represent molecular formulas of the losses. (a) A gap (−) is introduced for the missing CO loss in the left tree (dashed edge and node). Losses CO and CH3 are aligned by a mismatch (dotted edges). (b) In the left tree, the fragment after loosing H3N is missing (dashed edges and node), whereas the fragment after further loss of C2H2 is observed. To account for missing fragments, we introduce the join operation. It allows to align the two successive losses H3N and C2H2 in the right tree to a single loss C2H5N in the left tree (dotted edges). Fragments may be missing because the corresponding peak was not detected, for example